FIGURE 1.
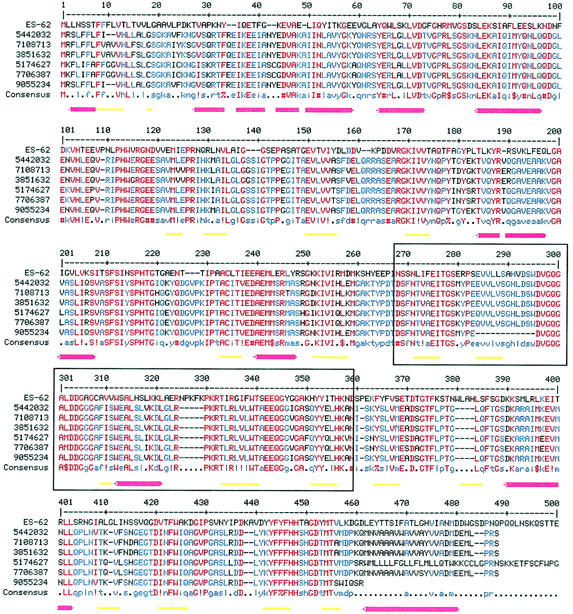
Multiple alignment of the amino acid sequence of ES-62 with those of its six protein homologs (Table 1). The multiple alignment was generated using MULTALIN (Corpet, 1988). The BLOSUM 62 matrix was used with a gap weight of 12 and a gap length weight of 2. The consensus levels were 90% for high homology (red) and 50% for low homology (blue). Symbols are as follows: ! is I or V; $ is L or M; % is F or Y; # is any one of NDQEBZ. The secondary structure for ES-62 predicted by JPRED (Cuff et al., 1998; Cuff and Barton, 1999) is shown beneath the alignment. Red cylinders represent α-helices and yellow arrows represent β-strands. In places helices appear broken reflecting the gaps inserted in the ES-62 sequence during alignment with its homologs. Boxed residues are those within ES-62 that are 32% identical with residues 74–168 of a leucyl aminopeptidase from Aeromonas proteolytica (PDB code, 1AMP).
