TABLE 2.
Elastic, geometric, and electrostatic parameters used in the DNA/protein model
| Parameter | Definition | Value |
|---|---|---|
| N | Number of the vertices in the circular DNA model | 176 (5.28 kbp) |
| l0 | Equilibrium segment length | 10 nm |
| le | Kuhn statistical length of DNA | 100 nm |
| k | Number of elastic segment for each Kuhn statistical length | 10 |
| C | Twisting rigidity constant | 3.0 × 10−19 erg·cm |
| kB | Boltzmann's constant | 1.38 × 10−23 J/K |
| T | Absolute temperature | 298 K |
| g | Bending rigidity constant of average DNA | pkBT/l0 |
| h | Stretching rigidity constant |  |
| Δt | Time step for Brownian dynamic simulations | 600 ps |
| d0 | Radial distance criterion for site juxtaposition | 10 nm |
| cS | Monovalent salt concentration | 0.2 M |
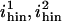 |
Vertices to which a Hin dimer can bind | 1, 34 |
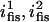 |
Vertices to which a Fis dimer can bind | 5, 6 |
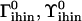 |
Equilibrium tilt-like and roll-like bends on an ihin vertex by a Hin dimer | −18°, 0° |
 |
Equilibrium tilt-like and roll-like bends on an ifis vertex by a Fis dimer | 60°, 0° |
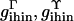 |
Computational tilt-like and roll-like bending rigidity for ihin vertices | g, g |
 |
Computational tilt-like and roll-like bending rigidity for ifis vertices | 3.0 g, 3.0 g |
| κ | Inverse Debye length (salt-dependent, here for 0.2 M) | 1.477 nm−1 |
| λ | Effective linear charge density of double helix (here for 0.2 M) | 40.9 e/nm |
| ε | Relative dielectric constant of aqueous medium | 80 |
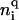 |
Number of effective charges for a DNA segment, a Hin-dimer/DNA complex, and a Fis-dimer/DNA complex, respectively | 5, 200, 100 |
| μ | Computational short-range repulsion force | 35 pN |
| ρ0 | Hydrodynamic radius of a DNA segment of l0 | 2.24 nm |
