TABLE 4.
Computation of the 〈3JNHα〉 coupling constant
| Peptide sequence* | Boltzmann-averaged† value of the vicinal coupling constant 〈3JNHα〉‡ | Zimmerman code of the lowest energy conformation§ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PPP¶ | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 4.6 | 8.6 | FFFFFFADD | |
| PPP∥ | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 5.1 | 4.9 | 8.8 | 6.6 | FFFFFFADA | |
| PPP** | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 5.3 | 3.4 | 3.7 | 6.9 | FFFFFFAAD | |
| PAP¶ | 3.4 | 3.4 | 5.3 | 6.6 | 5.2 | 5.3 | 5.3 | 4.7 | 8.6 | FFCDFFADD | |
| PAP∥ | 5.3 | 3.7 | 5.2 | 8.0 | 5.3 | 5.3 | 4.8 | 9.5 | 6.6 | FFADFFADA | |
| PAP** | 5.3 | 5.3 | 3.4 | 6.8 | 5.3 | 5.3 | 3.4 | 4.1 | 4.3 | FFFF*FFAAF | |
| PAAP¶ | 5.1 | 5.3 | 5.3 | 4.9 | 6.8 | 5.3 | 5.3 | 3.5 | 4.4 | 6.5 | FFAAFFFCD*D |
| PAAP∥ | 5.2 | 5.3 | 5.3 | 6.8 | 8.0 | 5.2 | 5.3 | 5.1 | 8.6 | 6.3 | FFAADFFADA |
| PAAP** | 5.3 | 3.4 | 5.3 | 6.8 | 6.8 | 5.3 | 5.3 | 3.4 | 4.2 | 6.2 | FFAA*A*FFACG |
| PQP¶ | 5.3 | 4.0 | 5.3 | 8.5 | 4.7 | 4.7 | 5.3 | 4.8 | 7.4 | FFAEFFAA*D | |
| PQP∥ | 5.3 | 3.7 | 5.0 | 8.6 | 5.3 | 5.3 | 5.0 | 9.1 | 6.6 | FFADFFADA | |
| PQP** | 5.3 | 5.3 | 3.4 | 6.8 | 3.4 | 3.4 | 3.4 | 5.1 | 4.4 | FFAA*FFFAF | |
| PGP¶ | 4.7 | 5.2 | 4.7 | 2.9 | 5.1 | 5.1 | 5.3 | 5.3 | 8.3 | FFFD*FFAGF | |
| PGP∥ | 5.1 | 4.3 | 4.9 | 7.5 | 5.2 | 5.2 | 4.7 | 9.4 | 6.8 | FFADFFADA | |
| PGP** | 3.4 | 3.4 | 3.4 | 6.2 | 5.3 | 5.3 | 3.4 | 2.9 | 7.9 | FFADFFAAA | |
| PVP¶ | 5.3 | 3.5 | 5.3 | 8.7 | 5.3 | 5.3 | 5.3 | 5.0 | 9.1 | FFCDFFADD | |
| PVP∥ | 5.2 | 4.8 | 4.3 | 9.6 | 4.7 | 5.3 | 4.4 | 7.9 | 6.3 | FFADFFADA | |
| PVP** | 3.4 | 5.3 | 3.4 | 5.4 | 3.4 | 5.3 | 5.3 | 6.0 | 7.6 | FFAAFFACA | |
PXP and PXXP represent the X residue in the sequence Ac-PPPXPPPGY-NH2 (for X = Pro, Ala, Gln, Gly, and Val) and Ac-PPPXXPPPGY-NH2 (for XX = AlaAla), respectively.
Values computed by using all the accepted conformations listed in Table 1. The values of the coupling constants of all the non-proline residues in the sequence are in boldface. The values for the X residue in the sequence PXP and PAAP are underlined.
The theoretical values of the coupling constants were computed from the calculated values of the dihedral angle φ by using the Karplus relation (Karplus, 1959, 1963): 3JNHα = A cos2ϕ − B cos ϕ + C, with ϕ = |φ − 60.0| and A = 6.4, B = 1.4, and C = 1.9, as parameterized by Pardi et al. (1984).
Conformations are classified in terms of the regions of the φ–ψ Ramachandran (Ramachandran et al., 1963) map in which they occur (Zimmerman et al., 1977). On the left-hand half of the map (φ < 0°), the regions are defined as A–G; on the right-hand half of the map (φ ≥ 0°), the regions are defined by inversion of the left-hand half around the center of the map and an asterisk is appended to the letters. The conformations of all non-proline residues in the sequence are in boldface. The X residue in the sequence PXP and PAAP is underlined.
The calculations were carried out by using a GP potential, as explained in Methods.
The calculations were carried out by using a GPSAS potential, as explained in Methods.
The calculations were carried out at pH 7 at t = 25°C by using the GPSP potential, as described in Methods. The value of 10.10 was adopted as the 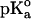 for the ionizable group of the residue Tyr, as an average from the data of Perrin (1972).
for the ionizable group of the residue Tyr, as an average from the data of Perrin (1972).
