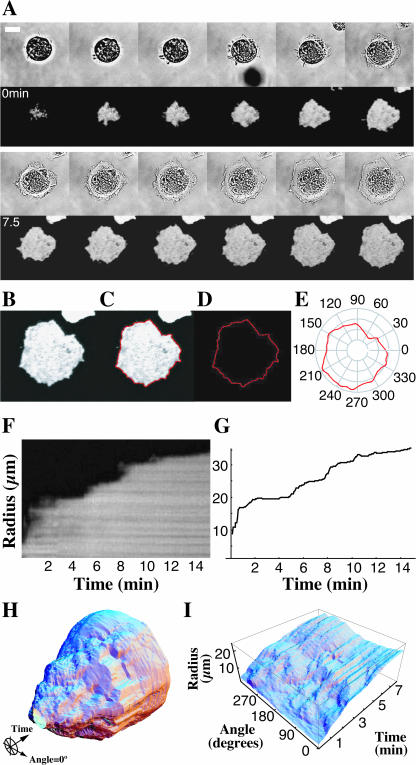
FIGURE 1.
Computer visualization of spreading in space and time. A time-lapse series of bright-field (A, upper series) and total internal reflection fluorescence (TIRF) microscope images (A, lower series) of a calcein AM-loaded fibroblast spreading from suspension onto a fibronectin-coated substrate (scale bar = 20 μm, 75 s between frames, 15 min total length). Individual TIRF images (B) were analyzed via a program that found the border between fluorescent cell and background (C and D) and then expressed this border in polar coordinates (E). F shows the traditional method for measuring motility via a kymograph that gives a plot of radius as a function of time along a single line perpendicular to the cell edge. Our method reproduces data in this single spatial dimension (G). Our technique allows us to easily visualize the time course of cell spreading in two spatial dimensions by displaying the entire sequence in cylindrical coordinates with radius as a function of angle and time (H). To view spreading of an entire cell at the same time, we express radius as a function of angle and time in Cartesian coordinates (I).