FIGURE 1.
Model-predicted FRAP recovery curves. The FRAP recovery predicted by each idealized model is compared to that predicted by the full model (Df = 30 μm2/s, w = 0.5 μm). Domains are identified where the full model is well fit by an idealized model (A–C), but this is not always the case (D). (A) With very weak binding, the pure-diffusion dominant model (red line) provides a good fit (sum of residuals = 0.08) to the full model (bold black line). Note that the effective diffusion model result is obscured by the pure-diffusion dominant curve and also provides a good fit; pure-diffusion dominant is a particular case of the effective diffusion model, namely Deff = Df. The reaction dominant model (orange line) provides a very poor fit. (B) With a high 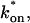 the effective diffusion model (green line) alone provides a reasonable fit to the full model (sum of residuals = 0.89). (C) With low
the effective diffusion model (green line) alone provides a reasonable fit to the full model (sum of residuals = 0.89). (C) With low 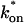 and low koff, the reaction dominant model (orange line) provides a good fit with the full model (sum of residuals = 0.66). (D) For certain values of
and low koff, the reaction dominant model (orange line) provides a good fit with the full model (sum of residuals = 0.66). (D) For certain values of 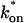 and koff, none of the idealized models result in a good fit to the full model (pure-diffusion dominant sum of residuals = 16.6; effective diffusion sum of residuals = 8.30; and reaction dominant sum of residuals = 8.54).
and koff, none of the idealized models result in a good fit to the full model (pure-diffusion dominant sum of residuals = 16.6; effective diffusion sum of residuals = 8.30; and reaction dominant sum of residuals = 8.54).

