FIGURE 1.
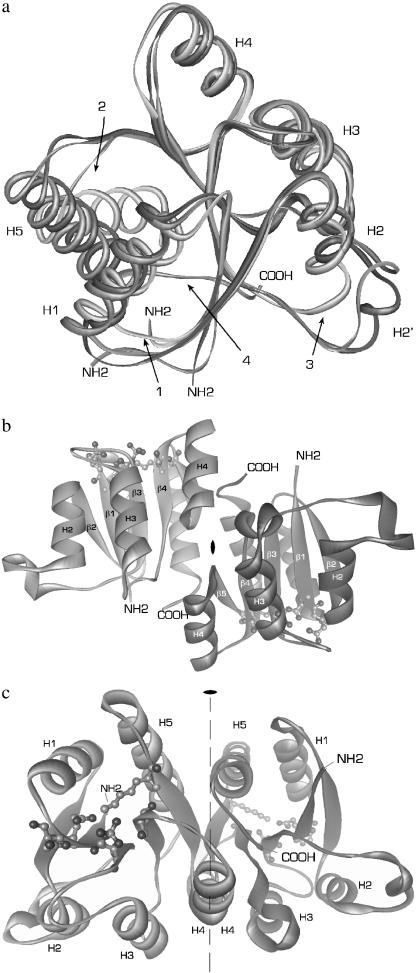
Overall structure of phospho-RcpA. (a) Overlaid structures of RcpA (dark gray), RcpB (gray), and CheY (light gray) illustrating the overall structural homology and the differences in (1) loop H1/β2, (2) loop β5/H5, and (3) loop H2/β3. In the structure of CheY, H5 is tilted with respect to H5 in RcpA and B. The extended C-termini of the cyanobacterial RRs (4), not present in CheY, participate in the dimer interaction. (b and c) For simplicity, the protein backbones are depicted as ribbons. The illustration shows dimeric phospho-RcpA as viewed from the (b) side and (c) top with the noncrystallographic twofold rotational axis. The active site in each subunit is drawn as ball and stick. The protein N- and C-termini are labeled. Phospho-RcpA crystallized in space group P21212 and diffracted to 1.9 Å. The final model comprises residues 10–148; residues 1–9 and 149 are not visible in the electron density, presumably due to disorder. Apo-RcpB (not shown) formed crystals in space group P41212 which diffracted to 1.6 Å (SeMet) and 1.75 Å (wild-type). The phase information obtained from the SeMet derivative was used to solve and refine the structure of wild-type apo-RcpB to 1.75 Å.