TABLE 1.
A set of parameter values that give a good fit between mean-field theory and the SB data for the six mutant strains
| m | 0 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|
| E0(1, m) | 0.864 | 0.310 | −0.147 | −8.02 | −20.1 |
| E0(2, m) | 4.69 | – | −1.04 | – | −7.97 |
| E1(1, m) | 30 | 2.10 | 1.41 | 0.763 | 0.740 |
| E1(2, m) | 30 | – | 30 | – | 30 |
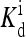 (1, m) (1, m) |
2.43 | 21.3 | 18.0 | 0.0314 | 6.49 × 10−7 |
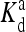 (1, m) (1, m) |
3.7 × 109 | 128.9 | 85.4 | 204.2 | 727.1 |
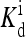 (2, m) (2, m) |
0.257 | – | 32539 | – | 24.9 |
 (2, m) (2, m) |
2.5 × 1010 | – | 9.8 × 1017 | – | 7.7 × 1017 |
|
EJ(q,q′)
|
q′ = 1
|
q′ = 2
|
|||
| q = 1 | 0 | −4.40 | |||
| q = 2 | −6.41 | −1.38 | |||
The local parameters for each receptor type, E0, E1 (in unit of thermal energy) and 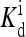 (in unit of μM), are listed for each relevant receptor type characterized by (q, m), where q = 1 for Tar and q = 2 for Tsr, and m is the receptor methylation level. Values of
(in unit of μM), are listed for each relevant receptor type characterized by (q, m), where q = 1 for Tar and q = 2 for Tsr, and m is the receptor methylation level. Values of 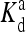 (q, m) =
(q, m) = 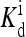 exp(E1 − E0) are also included for reference; the effective dissociation constant is a complicated combination of
exp(E1 − E0) are also included for reference; the effective dissociation constant is a complicated combination of  and
and 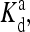 and it also depends on the actual activity. The coupling constants EJ(q, q′) (listed in the lower table) depend on the receptor species labels q and q′ for two neighboring receptors.
and it also depends on the actual activity. The coupling constants EJ(q, q′) (listed in the lower table) depend on the receptor species labels q and q′ for two neighboring receptors.
