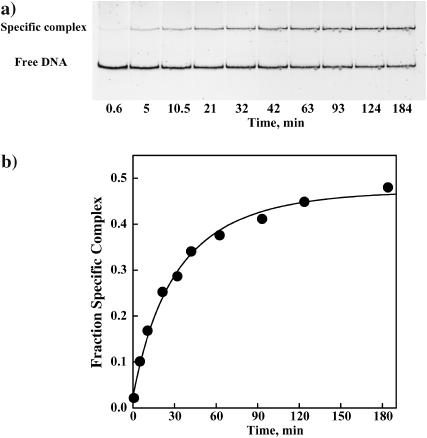
FIGURE 2.
(a) A polyacrylamide gel illustrating the kinetics of specific EcoRI binding to the 360-bp DNA fragment in the presence of the TAATTC “star” sequence oligonucleotide. In the absence of oligonucleotide, specific sequence binding is complete within 5 min. The dissociation rate of EcoRI from the TAATTC oligonucleotide complex is determined from the time dependence. The gel mobility shift assay is used to monitor the gain of specific EcoRI-DNA complex. EcoRI (1.5 nM) was incubated for different times with a mixture of specific sequence DNA fragment (3 nM) and the TAATTC sequence oligonucleotide. The particular conditions used in this experiment were 90 mM NaCl, 3.95 Osm betaine glycine, 20 mM imidazole (pH 7.0), and 20°C. The TAATTC sequence oligonucleotide was present in 250-fold molar excess over the specific sequence fragment. (b) The fraction of the specific EcoRI-DNA fragment complex is shown plotted against time for the gel shown in panel a. The solid line is the best fit of Eq. 7 to the data. The dissociation rate constant k4 for this complex was calculated as 2.4 min−1.