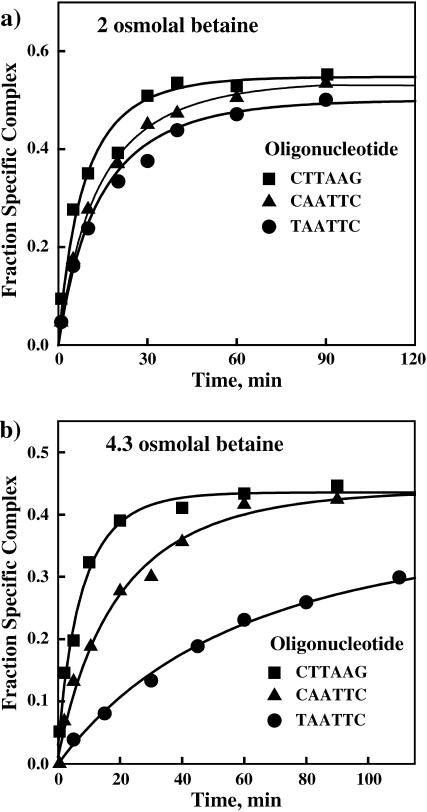
FIGURE 4.
Noncognate complex dissociation rates are strongly dependent on osmotic stress and the oligonucleotide sequence. Dissociation of EcoRI from the noncognate oligonucleotides is monitored by the increase in protein binding to the specific 360-bp DNA fragment with the incubation time. (a) In 2 Osm betaine glycine, the kinetic curves are similar for the three oligonucleotide complexes. All three noncognate oligonucleotides are present at 1000-fold molar excess over the specific DNA fragment. The salt and pH conditions used in this experiment were 90 mM NaCl, 20 mM imidazole (pH 7.0), 20°C. (b) At 4.3 Osm betaine glycine, the dissociation rates are clearly different for the three oligonucleotides. Both of the “star” sequence complexes dissociate substantially more slowly than the nonspecific inverted sequence complex. Of the “star” sequence complexes, the dissociation rate of the TAATTC oligonucleotide complex is significantly slower than that of the CAATTC oligonucleotide complex. All three noncognate oligonucleotides are present in 100-fold molar excess over specific DNA fragment.