TABLE 1.
Summary of threading results from PROSPECTOR_3 and final models by TASSER
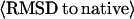 § §
|
Nfold¶
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| N* | Template selected† |
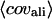 ‡ ‡
|
Tali | Mali | Mful | Tali | Mali | Mful | |
| Easy set | 593 (80%) | Top two plus consensus | 83% | 5.9 Å | 4.7 Å | 6.4 Å | 418 (363) | 481 | 396 (67%) |
| Medium set | 150 (20%) | Top five | 45% | 12.4 Å | 9.3 Å | 15.7 Å | 45 (2) | 71 | 12 (8%) |
| Hard set | 2 (0.3%) | Top 20 | 41% | 17.3 Å | 13.1 Å | 18.1 Å | 0 (0) | 0 | 0 (0%) |
| Single domain | 487 (65%) | 76% | 7.2 Å | 5.4 Å | 7.7 Å | 307 (258) | 377 | 296 (61%) | |
| Multiple domain | 258 (35%) | 73% | 7.4 Å | 6.1 Å | 9.5 Å | 156 (107) | 175 | 112 (43%) | |
| Membrane proteins | 18 (2%) | 71% | 10.7 Å | 7.5 Å | 12.0 Å | 8 (5) | 10 | 6 (33%) | |
| All | 745 | 75% | 7.2 Å | 5.6 Å | 8.3 Å | 463 (365) | 552 | 408 (55%) | |
Number of the target proteins in each category and the percentage in whole benchmark.
Number of templates used in the TASSER assembly procedure.
Average alignment coverage for the best template that has the lowest RMSD to native.
Average RMSD to native: Tali, the best template with RMSD calculated over aligned regions; Mali, the best model in top five with RMSD calculated over the same aligned regions as that in the threading template; and Mful, the best model in top five with RMSD calculated over entire chain.
Number of targets with RMSD to native below 6.5 Å: Tali, the best template with RMSD calculated over aligned regions. The numbers in parentheses are the templates of the alignment coverage ≥70%; Mali, the best model in top five with RMSD calculated over the same aligned regions as that in the threading template; and Mful, the best model in top five with RMSD calculated over entire chain. The value in parentheses is the fraction of targets in the specified category.
