TABLE 2.
TASSER modeling result for 1809 unaligned loop/tail regions of length greater than or equal to four residues
| Loops*
|
Tails†
|
|||
|---|---|---|---|---|
| RMSDglobal‡ | RMSDlocal§ | RMSDglobal‡ | RMSDlocal§ | |
| Total number¶ | 1345 (837) | 1345 (837) | 464 | 464 |
| NRMSD<1Å‖ | 55 (55) | 502 (400) | 3 | 46 |
| NRMSD<2Å | 167 (166) | 887 (617) | 12 | 133 |
| NRMSD<3Å | 338 (327) | 1119 (744) | 28 | 194 |
| NRMSD<4Å | 561 (497) | 1241 (804) | 50 | 246 |
| NRMSD<5Å | 810 (670) | 1280 (819) | 73 | 279 |
| NRMSD<6Å | 990 (773) | 1314 (830) | 99 | 304 |
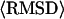 ** **
|
5.03 (4.03) Å | 1.82 (1.47) Å | 15.33 Å | 6.38 Å |
Result for unaligned loop regions. The data in parentheses is for the loops with RMSD of the stem residues below four Å.
Result for unaligned tail regions.
RMSD between native and the modeling loops (tails) after superposition of up to five neighboring stem residues on both sides (single side) of the modeling regions if applicable.
RMSD between native and the modeling loops/tails with direct superposition in the modeling regions.
Total number of the modeling regions.
Number of targets with a RMSD to native below the specific threshold values.
Average values of the RMSD to native for all unaligned loop/tail regions with length greater than or equal to four residues.
