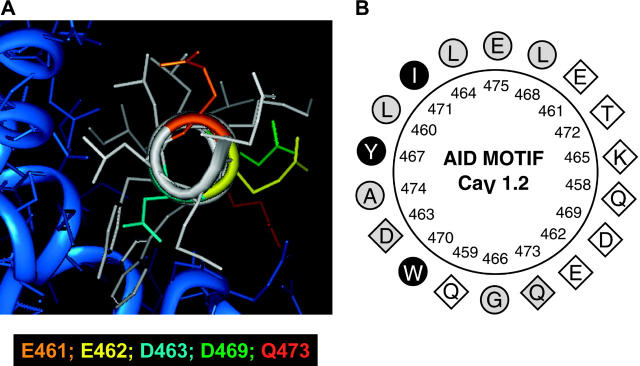
FIGURE 1.
(A) Three-dimensional representation of the AID helix of the rabbit CaV1.2 obtained with INSIGHT II using the human Cav1.2 crystal structure cocrystallized with CaVβ2a (Protein Data Bank: 1T0J.pdb) as a template (Van Petegem et al., 2004). The core of the AID helix appears in white. Side chains are color-coded: E461 (orange), E462 (yellow), D463 (turquoise), D469 (green), and Q473 (red). The hydrophobic face of the AID helix interacts with the CaVβ2a subunit shown in blue. (B) Two-dimensional helical wheel representation of the AID region between Q458 and E475 as predicted in the rabbit CaV1.2. Helical wheel projections were carried out with ANTHEPROT. As seen, the hydrophobic (circles) and hydrophilic (diamonds) residues line up on opposite sides of the helix. Filled black symbols highlight the residues that were shown to interact strongly with CaVβ2a or CaVβ3 in every crystal structure published to this date (Van Petegem et al., 2004; Chen et al., 2004; Opatowsky et al., 2004); the empty symbols represent residues that were shown to be clearly noninteracting with either CaVβ2a or CaVβ3; and the shaded symbols show residues for which some degree of interaction was found in either crystal structure.