TABLE 3.
Results of fits to 17O and 2H MRD data from SNase mutants at 20°C
| From 17O MRD data
|
From 2H MRD data
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SNase variant | pH | CP (mM)* | 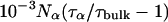 |
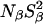 |
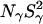 |
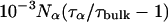 |
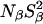 |
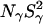 |
| Δ+PHS | 4.5 | 1.59 | 2.0 ± 0.1 | 1.45 ± 0.04 | 3.8 ± 0.3 | 1.5 ± 0.3 | 3.2 ± 0.1 | 4.8 ± 0.8 |
| Δ+PHS | 7.0 | 1.59 | 2.3 ± 0.2 | 1.70 ± 0.06 | 5.0 ± 0.4 | 2.1 ± 0.2 | 2.2 ± 0.1 | 4.6 ± 0.4 |
| Δ+PHS | 9.5 | 1.59 | 2.5 ± 0.2 | 1.73 ± 0.06 | 4.3 ± 0.4 | 1.9 ± 0.5 | 4.8 ± 0.2 | 12 ± 2 |
| Δ+PHS/V66K | 4.5 | 0.85 | 1.7 ± 0.2 | 1.64 ± 0.06 | 3.3 ± 0.5 | 1.4 ± 0.4 | 3.4 ± 0.2 | 3.9 ± 1.0 |
| Δ+PHS/V66K | 7.0 | 1.44 | 2.4 ± 0.2 | 1.76 ± 0.06 | 4.0 ± 0.5 | 2.1 ± 0.2 | 2.1 ± 0.1 | 4.3 ± 0.6 |
| Δ+PHS/V66E | 7.0 | 1.18 | 2.3 ± 0.2 | 1.81 ± 0.06 | 4.5 ± 0.4 | 2.2 ± 0.3 | 2.1 ± 0.1 | 4.5 ± 0.9 |
| Δ+PHS/V66E | 9.5 | 1.00 | 2.1 ± 0.2 | 1.81 ± 0.07 | 4.6 ± 0.5 | 1.5 ± 0.5 | 4.8 ± 0.2 | 14 ± 2 |
All MRD profiles were subjected to bi-Lorentzian fits with three adjustable parameters and the two correlation times were fixed at the mean values, τβ = 12.6 ns and τγ = 2.1 ns, obtained from unconstrained fits.
Protein concentration in MRD sample.
