TABLE 1.
Parameters determined from least-square fits to the hemoglobin MRD data
| A′ | B1 | τ1 | B2 | τ1 | |
|---|---|---|---|---|---|
| Formate | |||||
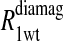
|
0.02 | 2.8E + 04 | 7.6E − 08 | 1.8E + 06 | 1.2E − 09 |
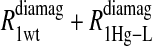
|
0.01 | 7.7E + 04 | 1.2E − 07 | 1.1E + 06 | 2.1E − 09 |
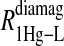
|
0.00 | 5.8E + 04 | 1.3E − 07 | ||
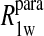
|
−0.27 | 3.4E + 09 | 2.9E − 11 | 3.0E + 07 | 1.7E − 09 |
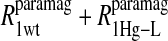
|
−0.23 | 3.3E + 09 | 3.2E − 11 | 4.0E + 07 | 1.9E − 09 |
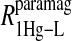
|
0.02 | 1.6E + 08 | 5.3E − 11 | 1.1E + 07 | 2.1E − 09 |
| Acetate | |||||
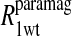
|
−0.03 | 3.5E + 08 | 3.4E − 11 | 4.3E + 06 | 4.2E − 09 |
| Water | |||||
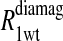
|
0.02 | 4.5E + 04 | 5.4E − 08 | 2.0E + 06 | 8.8E − 10 |
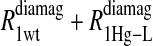
|
0.02 | 4.6E + 04 | 6.2E − 08 | 1.8E + 06 | 1.1E − 09 |
Eq. 4 with N = 2 was applied to the 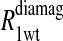 and the
and the 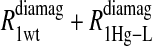 data for both water and formate protons. The constant A′ represents the residual relaxation rate constant remaining at the high field end for all of the fits tabulated here. The data for
data for both water and formate protons. The constant A′ represents the residual relaxation rate constant remaining at the high field end for all of the fits tabulated here. The data for 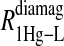 of the formate proton was fit in a similar fashion except with N = 1. Eq. 3 with N = 2 and physical constants corresponding to an electron was applied to the
of the formate proton was fit in a similar fashion except with N = 1. Eq. 3 with N = 2 and physical constants corresponding to an electron was applied to the 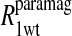 and the
and the 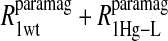 data for the formate protons and to the
data for the formate protons and to the 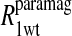 data for the acetate protons. The units of the correlation times are in seconds (s) and for the relaxation rates are s−1. Errors for the fitted parameters are estimated at 20%.
data for the acetate protons. The units of the correlation times are in seconds (s) and for the relaxation rates are s−1. Errors for the fitted parameters are estimated at 20%.
