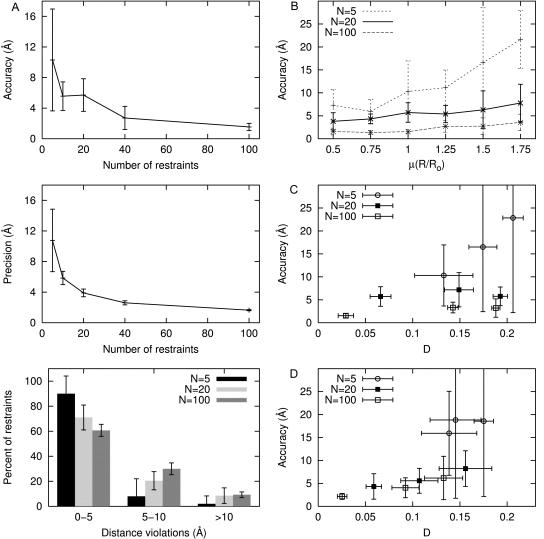
FIGURE 3.
Distance-restrained docking: benchmarking. (A) Sensitivity to number of restraints: (top) accuracy (defined as the distance between the maximum likelihood model and the target), (middle) precision (defined as the mean distance between the maximum likelihood model and all other accepted models), and (bottom) distribution of distance violations for models generated from simulated FRET data with flexibly tethered probe sites uniformly distributed about the fixed chromophore target, μ(R/Ro) = 1 and 15% random error. (B) Sensitivity to distance distribution of restraints. Models generated from simulated FRET data with flexibly tethered probe sites symmetrically distributed about the fixed target and 15% random error while varying μ(R/Ro). (C) Sensitivity radial distribution of restraints. Models generated from simulated FRET data with μ(R/Ro) = 1, 15% random error, and flexibly tethered probe sites located in one octant (large D), located in one quadrant (intermediate D), or distributed symmetrically (small D) about the fixed target. (Because results for models with chromophore sites located in one hemisphere or distributed symmetrically are similar to one another, the latter are omitted for clarity.) (D) Sensitivity to systematic error in restraints. Models generated from simulated FRET data with μ(R/Ro) = 1.0, 15% random error, 10% systematic error, and flexibly tethered probe sites located in one quadrant (large D), located in one hemisphere (intermediate D), or distributed symmetrically (small D) about the fixed target. (Because results for models with chromophore sites located in one octant or one quadrant are similar, the latter are omitted for clarity.)