FIGURE 3.
Parameters characterizing confined diffusion are severely affected by time-averaging over the frame time. (A) The MSD-t plots for particles trapped within a 120-nm length square compartment at frame times of 25 μs (left) and 33 ms (right). MSDs calculated from the trajectories shown in Fig. 2 (single-molecule MSDs), plotted as a function of the time interval. MSD grows rapidly and then levels off. The theoretical plateau value is 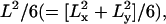 which is 4800 nm2 (because both Lx and Ly are 120 nm). At a frame time of 25 μs (left figure), this is very close to the observed value. However, at a frame time of 33 ms, the limiting plateau value is severely suppressed (right figure; note the greatly reduced value in the y axis, despite the expanded x axis from the left figure by 1000-fold), corresponding to the centralized distribution of the observed points at this frame time. (B) Apparent microscopic diffusion coefficient D2–4, plotted against frame time. The solid line is not a fitting, but the predicted apparent diffusion coefficient from Eq. 1 (see the text). The set diffusion coefficient in the simulation was 9 μm2/s. (C) The compartment size determined by fitting the MSD-t curve (like those shown in A) using Eq. 2 displayed as a function of the frame time (at least 100 simulations at each frame time). Note that both the apparent diffusion coefficient and compartment size are severely underestimated at longer frame rates, as expected from the trajectories in Fig. 2 and the MSD-t curves in A. In B and C, error bars are mostly hidden by the data markers.
which is 4800 nm2 (because both Lx and Ly are 120 nm). At a frame time of 25 μs (left figure), this is very close to the observed value. However, at a frame time of 33 ms, the limiting plateau value is severely suppressed (right figure; note the greatly reduced value in the y axis, despite the expanded x axis from the left figure by 1000-fold), corresponding to the centralized distribution of the observed points at this frame time. (B) Apparent microscopic diffusion coefficient D2–4, plotted against frame time. The solid line is not a fitting, but the predicted apparent diffusion coefficient from Eq. 1 (see the text). The set diffusion coefficient in the simulation was 9 μm2/s. (C) The compartment size determined by fitting the MSD-t curve (like those shown in A) using Eq. 2 displayed as a function of the frame time (at least 100 simulations at each frame time). Note that both the apparent diffusion coefficient and compartment size are severely underestimated at longer frame rates, as expected from the trajectories in Fig. 2 and the MSD-t curves in A. In B and C, error bars are mostly hidden by the data markers.

