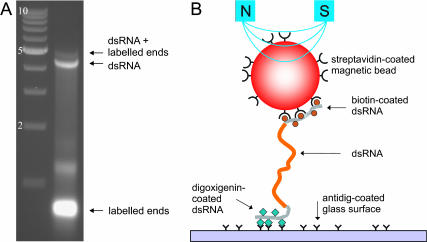
FIGURE 1.
(A) Gel electrophoresis analysis (nondenaturing agarose, 0.75%) of a dsRNA construct before placement in the magnetic tweezers. The left lane shows a reference ladder of dsDNA fragments at 1-kb intervals, with the qualification that on gel dsRNA is known to migrate slightly slower than dsDNA. The right lane shows four bands. The two most rapidly moving bands correspond to the labeled fragments (with the vast majority corresponding to 0.4-kb labeled fragments and a small minority corresponding to blunt-ended ligated dimers thereof). The slowly moving bands (near 4 kb on the reference ladder) correspond to the 4.2 kb dsRNA molecule without the labeled fragments and the 4.2 kb dsRNA molecule ligated to 0.4 kb biotin- and digoxigenin-labeled ends, respectively. (B) Cartoon of a dsRNA molecule as placed in the magnetic tweezers (not to scale). The dsRNA molecule is tethered to a magnetic bead by a biotin-streptavidin linkage and to the bottom glass surface of a flow cell using an antidigoxigenin-digoxigenin linkage. Magnets are placed above the flow cell to exert a force on the magnetic bead, and hence on the dsRNA molecule. In the experiments, the main part of the dsRNA molecule consists of 4.2 kb or 8.3 kb (∼1 μm or ∼ 2 μm), whereas the labeled ends each have a length of 0.4 kb or ∼ 0.1 μm.