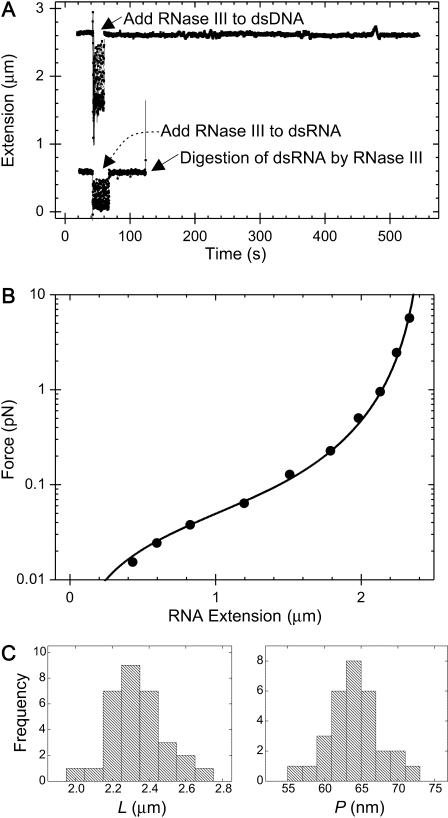
FIGURE 2.
(A, top) Extension of a dsDNA molecule is tracked in real time using magnetic tweezers. At the point indicated by the arrow, RNase III is added to the sample. Degradation of the molecule by RNase III would result in an upward movement of the magnetic bead and hence a disruption of the tracking of the extension of the molecule. Since this is not observed, we can conclude that the molecule does not contain any dsRNA, as expected. (Bottom) Extension of a dsRNA molecule is tracked in real time using magnetic tweezers. As before, RNase III is added to the flow cell. In this case, at time t∼ 130 s, the tracking of molecule's extension abruptly halts, indicating cleavage of a dsRNA molecule. (B) Example of a force-extension curve for 8.3 kb dsRNA (circles correspond to data points and the smooth curve corresponds to a fit to the wormlike chain model; Bouchiat et al., 1999). The fit yields a value of 63.0 ± 2.5 nm for the persistence length of this dsRNA molecule. (C) Histograms of the contour lengths (left) and the persistence lengths (right) measured for 31 8.3 kb dsRNA molecules. The mean contour length 〈L〉 measured was 2.33 ± 0.03 μm (N = 31) and the mean persistence length 〈P〉 measured was 63.8 ± 0.7 nm (N = 31).