TABLE 2.
Fluorescence anisotropy parameters for 10 nM TR12-aptamer-IgE binding (25°C)
| [IgE]*†‡ | φ1 (ns) | β1 | φ2 (ns) | β2 | φ3 (ns) | β3 | χ2 |
|---|---|---|---|---|---|---|---|
| 0 nM | 0.5 ± 0.2 | 0.28 ± 0.04 | 5.0 ± 0.4 | 0.72 ± 0.04 | – | – | 1.91 |
| 5 nM | 0.5 ± 0.2 | 0.26 ± 0.06 |  |
0.61 ± 0.05 | 50§ | 0.14 ± 0.07 | 1.47 |
| 10 nM | 0.7 ± 0.3 | 0.26 ± 0.11 | 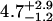 |
0.50 ± 0.08 | 50§ | 0.24 ± 0.13 | 1.91 |
| 15 nM | 1.0 ± 0.5 | 0.25 ± 0.16 | 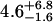 |
0.39 ± 0.13 | 50§ | 0.36 ± 0.19 | 1.43 |
| 26 nM | 0.5 ± 0.3 | 0.17 ± 0.12 | 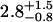 |
0.28 ± 0.07 | 50§ | 0.54 ± 0.14 | 1.74 |
| 150 nM | 0.7 ± 0.3 | 0.23 ± 0.07 | 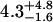 |
0.28 ± 0.05 | 50§ | 0.49 ± 0.09 | 1.55 |
All solutions are in binding buffer.
Samples are excited at 590 nm and emission is collected at 612 nm.
Anisotropy fitting function is: r(t) = r0 Σ βi exp(−t/φi).
This rotational correlation time is fixed to account for overall protein rotation.
