TABLE 1.
Static free energies from PB/LRA and MDFE
| Residue* | State | 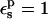 |
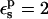 |
MDFE† |
|---|---|---|---|---|
| ASP | 95.5(0.3) | 116.8(0.3) | 124.2(2.2) | |
| Thioredoxin Asp-26 | Midpoint | 50.5(0.7) | 59.5(0.4) | 44.8(2.0) |
| ASPH | 28.7(0.4) | 10.7(0.3) | −16.6(4.6) | |
| ASP | 130.0(0.8) | 136.1(0.5) | 142.3(2.4) | |
| RNase A Asp-14 | Midpoint | 70.0(0.7) | 67.4(0.3) | 56.8(3.2) |
| ASPH | 11.5(1.0) | 0.3(0.6) | −17.9(4.2) | |
| ASP | 131.9(0.5) | 136.8(0.2) | 143.5(0.8) | |
| Thioredoxin Asp-20 | Midpoint | 60.5(0.3) | 63.0(0.1) | 57.0(1.2) |
| ASPH | −9.3(0.3) | −9.6(0.1) | −19.0(0.4) | |
| ASP | 139.6(0.4) | 143.3(0.2) | 144.6(1.4) | |
| Model‡ | Midpoint | 66.4(0.1) | 67.7(0.1) | 58.4(0.8) |
| ASPH | −6.0(0.2) | −7.8(0.1) | −19.3(1.4) |
Free energies in kcal/mol. The signs correspond to the ASP → ASPH direction (protonation). The protein dielectric constant is 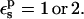 Mean values over 100–200 MD structures are reported. The statistical uncertainty of the PB static free energies (in parentheses) was determined by the method of Flyvbjerg and Petersen (1989). A correction has been added to the static terms to permit comparison with the MDFE derivatives (see text; it is, respectively, +2.0, −1.0, and −4.1 kcal/mol at the protonated (ASPH), midpoint, and charged (ASP) end state).
Mean values over 100–200 MD structures are reported. The statistical uncertainty of the PB static free energies (in parentheses) was determined by the method of Flyvbjerg and Petersen (1989). A correction has been added to the static terms to permit comparison with the MDFE derivatives (see text; it is, respectively, +2.0, −1.0, and −4.1 kcal/mol at the protonated (ASPH), midpoint, and charged (ASP) end state).
Free-energy derivatives from a molecular dynamics free-energy simulation (MDFE) starting from the ionized state ASP (“backward” run); see Table 1 in (Simonson et al., 2004).
The model compound (Fig. 1) is an aspartic acid with N-acetyl and N-methylamide blocking groups.
