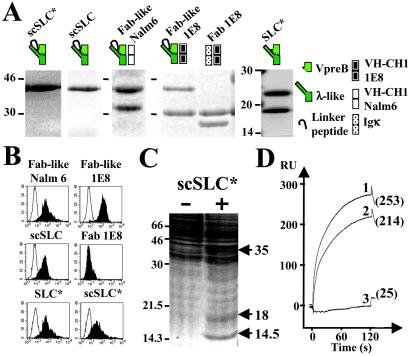
Figure 1.
Binding of soluble pre-BCRs or SLC components to MS5.1 cell line and biochemical identification of GAL1 as the SLC ligand. (A) Purified recombinant proteins were separated on SDS/15% polyacrylamide gels under reducing conditions and visualized by Coomassie blue staining. scSLC* (λ-like is fused to VpreB by a linker peptide) and SLC* (λ-like is reassociated with VpreB) were produced in E. coli and recombinant scSLC, Fab-like Nalm6 (scSLC+Nalm6 VH-CH1μ), Fab-like 1E8 (scSLC + 1E8 VH-CH1μ), and Fab 1E8 (κ+1E8 VH-CH1μ) by using baculoviruses. Schematic representations of the recombinant proteins are shown on the top of the figure. (B) Binding of the two Fab-like, Fab 1E8 and the different SLC recombinant proteins to MS5.1 cell line, were analyzed by flow cytometry. For each analysis, 10 μg/ml of recombinant proteins were used, and bindings were revealed by incubation with the 4G7 (2.5 μg/ml) anti-VpreB mAb (γ1, κ) for E. coli-derived proteins or with the M2 (5 μg/ml) anti-Flag mAb (γ1, κ) for baculovirus-derived proteins (black). For negative controls, the recombinant proteins are revealed with the HH5 (5 μg/ml) anti-HEL mAb (γ1, κ) (white). (C) Preparative MS5.1 cell lysate was incubated with nickel-Sepharose scSLC-loaded (+) and unloaded (−) beads (1.5 × 108 MS5.1 cells were used for each experimental condition). Material eluted in 6 M urea was separated on a preparative SDS/17.5% PAGE and revealed by Coomassie staining. Molecular weights of standard proteins are indicated in kDa. Arrows indicate differentially captured proteins. (D) BIAcore analysis of scSLC* (1), λ-like (2), and VpreB (3) binding to immobilized GAL1. The three analytes (40 μl at 10 μg/ml) were injected at a flow rate of 20 μl/min in HBS buffer on a dextran layer containing 500 resonance units (RU) of GAL1. The resulting sensorgrams are superimposed and are representative of two independent experiments. RU values at 125 s after the injection start are indicated.