Abstract
The genotypes of 63 strains (11 reference strains and 52 strains from hospitalized patients) of the haploid yeast Candida glabrata were determined from 33 putative gene enzymatic loci. This enabled the characterization of 26 different multilocus genotypes. Genetic differentiation was found between distant hospitals (located in Montpellier and Paris, France) but not for other parameters (anatomic origins or human immunodeficiency virus-positive [HIV+] and HIV− patients). Strong nonrandom association between loci could be seen. Such statistical linkages were confirmed upon comparing the patterns of 14 RAPD [random(ly) amplified polymorphic DNA] primers from 20 of these strains to results obtained from multilocus enzyme electrophoresis analysis. This finding suggests a mainly clonal mode of reproduction of C. glabrata. The consequences of the clonality displayed by C. glabrata populations on the epidemiology of this yeast are also discussed.
Analysis of the population genetics structure of natural populations elucidates the functioning and evolutionary capabilities of natural populations. It is particularly appropriate for organisms that cannot be directly observed (37). Indeed, the genotypic structure can provide useful clues for estimating the reproductive mode, gene flow, geographical structure, and approximate sizes of reproductive units (10).
Infectious diseases involve organisms whose population biology is difficult to assess. This is why molecular markers are useful tools in increasing our understanding of their epidemiology (40).
Candidoses are cosmopolitan infections due to yeast-like fungi. These yeasts are responsible for severe mucocutaneous or systemic infections, particularly in premature infants, pregnant women (in that case the infection is mildly self-limiting), after invasive intervention, in AIDS and immunodeficient patients (e.g., cancer, graft, and transplantation patients), and during chemotherapy (corticoids or antimitotics) and radiotherapy (26). Oropharyngeal candidosis is the earliest and the most common fungal infection found in AIDS patients (5, 6, 16). Moreover, susceptibility to other mycoses increases with the development of AIDS (17). Primary infections are essentially due to Candida albicans. After antifungal treatments, relapses are common and are often due to Candida glabrata (also known as Torulopsis glabrata) (2, 7, 18). Furthermore, the use of antifungal azoles seems to enhance the development of such mycoses (8, 15).
C. glabrata is a commensal yeast commonly isolated from the human digestive tract. This yeast is considered to be haploid and without a sexual cycle (45). Until know, very little attention has been devoted to the genetics of this opportunistic pathogen compared to the related C. albicans (1, 3, 28, 48). Thus, basic information on the biology and epidemiology of C. glabrata is needed.
We report here the enzyme polymorphism observed for 33 putative gene loci in 63 C. glabrata strains in 11 reference strains and in 52 strains isolated from hospitalized patients (22 in Paris and 30 in Montpellier). Furthermore, the patterns of 24 RAPD [random(ly) amplified polymorphic DNA] primers from 20 of these strains are compared to the multilocus enzyme electrophoresis (MLEE) results. This allows inferences to be made regarding the genetic structure and reproductive modality displayed by C. glabrata populations with respect to geography, infected organs, and the human immunodeficiency virus (HIV) status of the patient.
MATERIALS AND METHODS
Origin of the different strains.
We used 11 reference strains. Strain 774 comes from the Centraalbureau voor Schimmelcultures (Delft, The Netherlands), along with the reference CBS 138T; strains 918, 949, 957, 960, 962, 965, 970, and 971 were provided by the Institut Pasteur (Paris, France) with reference numbers IP 3, 811, 13, 20, 14, 12, 32, and 9, respectively, and strains 1109 and 1110 came from SANOFI (Montpellier, France) with reference numbers SANOFI 2223 and 2231. Information concerning the hospital strains (anatomic location, geographical origin, antifungal resistance, prophylaxy, and HIV+ versus HIV− status) is presented in Table 1. All strains were identified by the ID32 C (bioMérieux SA, Marcy l'Etoile, France) protocol. These strains were isolated, one per person, from 52 patients.
TABLE 1.
Sources of clinical isolates of C. glabrata strainsa
| Geographic origin (n), patient source, and isolate no. | Body site | Treatment |
|---|---|---|
| Paris (22) | ||
| HIV+ patients | ||
| 6032 | BL | N |
| 6034 | S | N |
| 6037 | S | N |
| 6039 | S | N |
| 6048 | S | F |
| 6049 | S | F+A |
| 6051 | S | N |
| 6052 | BC | N |
| 6054 | BL | N |
| 6055 | S | F+A |
| 6056 | S | N |
| HIV− patients | ||
| 6033 | U | N |
| 6035 | U | N |
| 6038 | V | N |
| 6041 | P | N |
| 6042 | BT | N |
| 6043 | BL | N |
| 6044 | BT | N |
| 6045 | BT | N |
| 6046 | P | N |
| 6047 | V | N |
| 6050 | V | N |
| Montpellier (30) | ||
| HIV+ patients | ||
| 1328 | BC | N |
| 1346 | BC | F |
| 1379 | BC | N |
| 1380 | BC | N |
| 1414 | BC | N |
| 1415 | BC | N |
| 1416 | BC | N |
| 1464 | BC | N |
| 1471 | BC | N |
| 1496 | BC | N |
| 1504 | BC | N |
| 1510 | BC | N |
| 1539 | BC | N |
| 1544 | BC | (F+K+I)b |
| 1545 | BC | (I)b |
| 1594 | BC | (K+I)b |
| 1595 | BC | (F+K+I)b |
| 1596 | BC | (F+K+I)b |
| 1599 | BC | N |
| 1600 | BC | N |
| 1614 | BC | N |
| 1616 | BC | N |
| 1617 | BC | N |
| 1619 | BC | N |
| 1620 | BC | N |
| 1622 | BC | N |
| 1651 | BC | N |
| 1656 | BC | N |
| HIV−patients | ND | N |
| 1291 |
Geographical origins: Hôpital Louis Mourier, Unité de Parasitologie, Mycologie, Colombes (Paris); Hôpital Gui de Chauliac, Clinique des Maladies Infectieuses A (Montpellier). n, Number of isolates. Body sites: B L, bronchioalveolar liquid; S, stool; B C, buccal cavity; U, urine; V, vagina; P, pharynx; BT, bronchial tubes; ND, not determined. Treatments: N, no treatment; F, fluconazole; A, amphotericin; K, ketoconazole; I, itraconazole.
Strain was resistant to the specified treatment.
Enzymatic and RAPD protocols.
Enzymatic extracts were obtained as previously described (28). Starch gel electrophoresis and enzymatic assays were performed as described previously (27, 33). Activity data were obtained for the following 26 enzymes: aldolase (EC 4.1.2.13), alpha-glyceraldehyde dehydrogenase (EC 1.1.1.8), creatine kinase (EC 2.7.3.2), diaphorase (EC 1.6.4.3), alcohol dehydrogenase (EC 1.1.1.1), esterase (EC 3.1.1.1), fructokinase (EC 2.7.1.4), fumarase (EC 4.2.1.2), glucose phosphate isomerase (EC 5.3.1.9), glucose-6-phosphate dehydrogenase (EC 1.1.1.49), isocitrate dehydrogenase (1.1.1.42), leucine aminopeptidase (EC 3.4.11), lactate dehydrogenase (EC 1.1.1.27), malic enzyme (EC 1.1.1.40), mannose-6-phosphate isomerase (EC 5.3.1.8), purine nucleoside phosphorylase (EC 2.4.2.1), peptidase A (EC 3.4.13, substrate Val-Leu), peptidase B (EC 3.4.13, substrate Leu-Gly-Gly), peptidase C (EC 3.4.13, substrate Lys-Leu), peptidase D (EC 3.4.13 substrate Phe-Pro), phosphoglucomutase (EC 2.7.5.1), glyceraldehyde phosphate dehydrogenase (EC 1.2.1.12), octopine dehydrogenase (EC 1.5.1.11), acid phosphatase (EC 3.1.3.2), sorbitol dehydrogenase (EC 1.1.1.14), and superoxide dismutase (EC 1.15.1.1.).
Creatine kinase, fructokinase, leucine aminopeptidase, mannose-6-phosphate isomerase, peptidase 3, phosphoglucomutase, and superoxide dismutase enzymatic activities were each expressed by two loci. Thus, data were obtained for 33 genetic loci. Alleles were numbered according to their anodal mobility.
To test for the possible correlation between two independent sets of genetic markers (MLEE and RAPD), a subset of 20 of the available strains was selected. These strains were chosen in order to examine the existing enzymatic variability (see below). Each strain was cultured in two vials containing Sabouraud agar (Difco) for 48 h at 27°C. Cultures were then suspended and ground in a cell homogenizer under CO2 monitored cold conditions (dry ice). DNA was extracted according to a standard protocol (19). Forty primers belonging to the E and F kits (Operon Technologies, Inc., Alameda, Calif.) were tested. A band was considered polymorphic if it is present (amplified) in at least two strains and not all strains. Among these primers we retained 24 bands (assumed to be loci) that were reproductive (one individual always displayed the same profile after each PCR). These 24 bands were obtained by using 14 primers (E4, E6, E14, E17, E18, F1, F2, F4, F6, F10, F12, F13, F14, and F16). Thus, one primer provides one or more bands that are each interpreted as a locus with two alleles (band present = 1, band absent = 0). We used this information for the comparative analysis between MLEE and RAPD data.
Data analysis.
The genetic distance used to build a dendrogram linking the different strains of C. glabrata was the Cavalli-Sforza and Edwards (4) chord distance matrix, which is the most appropriate for tree construction (38). The distances were computed by the GENETIX v.4 software package (Laboratoire Génome et Populations, CNRS UPR 9060, Université de Montpellier II, Montpellier, France). The distance matrix obtained was then used to build a dendrogram (neighbor-joining method) (35) computed by the software NJTree v2.0 of the RESTSITE v1.1 package (24). This dendrogram provided a visual picture to illustrate the genetic structure of our sample.
Linkage disequilibrium between loci gives a clue regarding the reproductive regime of the population under investigation.
Nonrandom association between each pair of loci was tested by the exact test for genotypic disequilibrium provided by the software GENEPOP v3.2.a (30). For each locus pair, an unbiased estimate of the P value of the probability test (or Fisher exact test) was operated by the Markov chain method (30). The total number of iterations (randomization) was set to 106. The probability of each randomized table is computed, and the P value is calculated as the sum of the probabilities of all tables (with marginal values the same as the observed one) with a probability lower than or equal to that of the observed table.
Additionally, the level of significance for nonrandom association between multilocus repeated genotypes was tested by the combinatorial probability of sampling a given genotype as often as or more often than that actually observed (d1) (41). A multilocus standardized index of linkage disequilibrium IsA was also computed by using LIAN 3.0 software (12), which tests the null hypothesis of no linkage by a Monte Carlo simulation (10,000 permutations) on the variance of genetic distances between isolates (VD) (12, 13).
The significance of genetic differentiation between strains obtained at the two different hospitals, between HIV+ and HIV− patients, or between strains found in the respiratory tract and those obtained in the anal or urogenital spheres was tested by the G-based exact test for population differentiation (11). The test is performed after 15,000 permutations of genotypes between samples by the software F-Stat 2.9 (http://www.unil.ch/izea/softwares/fstat.html) (9). For each permutation, the log-likelihood statistic (G) is computed, at each locus, from the allelic contingency table among populations. The P value corresponds to the proportion of randomized G that was above or equal to the G of the observed data. This software was also used to compute Fst (a standardized measure of genetic differentiation) unbiased estimates (42) for each locus and over all loci. The Fst value varies between 0 (no differentiation) and 1 (all samples fixed for a different allele).
Nei's 1972 genetic distance (21) was used for the study of correlation between distance matrices because it best estimates the branch length for electrophoretic data (38). The distances were computed by the GENETIX v.4 software package.
Allelic frequencies cannot be computed for RAPD markers. For these markers, the measure of genetic distances between strains was computed with Nei and Li's distance finding (23). The RAPDistance Package of Armstrong and coworkers (available from the Research School of Biological Sciences, Canberra, ACT 2601, Australia [http://life.anu.edu.au/molecular/software/RAPDistance]) was used. The correlation between the two half matrices obtained (MLEE and RAPD) was then tested by a Mantel test (20), which is appropriate for matrix comparisons. A mixture of individuals from differentiated population may generate linkage disequilibria (Wahlund effect). In order to control for the possible influence of geographical distances, Mantel tests were also made between the matrix of geographical distances coded 0 (local), 1 (Paris-Delft), 2 (Paris-Montpellier), and 3 (Montpellier-Delft) and genetic distances (MLEE or RAPD). Furthermore, each genetic distance matrix (MLEE and RAPD) was regressed against these geographical distances, and the residuals were kept. Theoretically, the residuals represent the part of the variance not explained by geographical distances. The matrix of these residuals was used for an additional Mantel test, which hopefully corrected for geographical influences. Mantel tests were performed by using GENEPOP v3.2.a.
Since multiple testing enhances type I error, we applied the sequential Bonferroni procedure when required (32). For example, if one kind of test is repeated 100 times on a population that fulfills the null hypothesis, the definition of statistical inferences predict that five of these tests will be significant at the 5% level. A technique to avoid this caveat is the sequential Bonferroni procedure, where the desired significant level (say, α) is divided by the number of remaining tests. Thus, for n tests, the lowest P value (among the n available) is compared to the corrected level α/n, the second lowest P value is compared to α/(n − 1), etc. The sequential Bonferroni significant P values will then be the i + 1 ones that stay below the corresponding corrected significant level, α/(n − i). As a complement, the proportion of tests significant at the 5% level was compared to the expected 0.05 proportion by an exact binomial test performed by using S-Plus 2000 (Professional Release 2; MathSoft, Inc.). This alternative procedure allows testing of whether the proportion of significant tests is equal to or below the 5% expected under the null hypothesis (at the 5% level of significance). This approach may be useful for procedures involving many tests that are not very powerful individually (small sample sizes) as in linkage disequilibrium testing between pairs of loci. In such cases, indeed, the sequential Bonferroni procedure may be too conservative.
The power of the tests was evaluated by running simulations of asexual clonal haploids with the software EASYPOP v1.6 (IZEA; Lausanne University, Lausanne, Switzerland [http://www.unil.ch/izea/softwares/easypop.html]). The computer simulation was performed with an island model (47) of 100 subpopulations of 100 haploid individuals each, with a migration rate of 0.04. The 33 loci (as in our data) displayed a random mutation rate of 10−5 into five possible allelic states. All of these parameters were found after a trial-and-error process involving several simulations with different parameter sets until we observed equilibrium values similar to those observed in our real samples in terms of the mean number of polymorphic loci (13.8 versus 13 in the real samples), unbiased heterozygosity (0.088 versus 0.11), and differentiation between samples (Fst = 0.12 versus 0.11). Each population began in a monomorphic state with a strict clonal mode of reproduction. The simulation ran for 10,000 generations (sufficient to reach a stable equilibrium between drift, migration, and mutation), after which 30 samples of 22 and 30 individuals (from Paris and Montpellier, respectively) were randomly sampled from 2 of the 100 subpopulations. These simulations provided a null hypothesis for a case of 100% clonality. In other words, this supplied 30 data sets equivalent to the C. glabrata samples we disposed of but with a 100% clonal mode of reproduction. Such data sets allowed us to test the power of detection of linkage disequilibria in sample sizes of 22 and 30 isolates drawn from a strictly asexual species in order to compared them to what we observed in C. glabrata isolates from Paris and Montpellier, where the reproductive mode is unknown.
RESULTS
Genetic variability within C. glabrata.
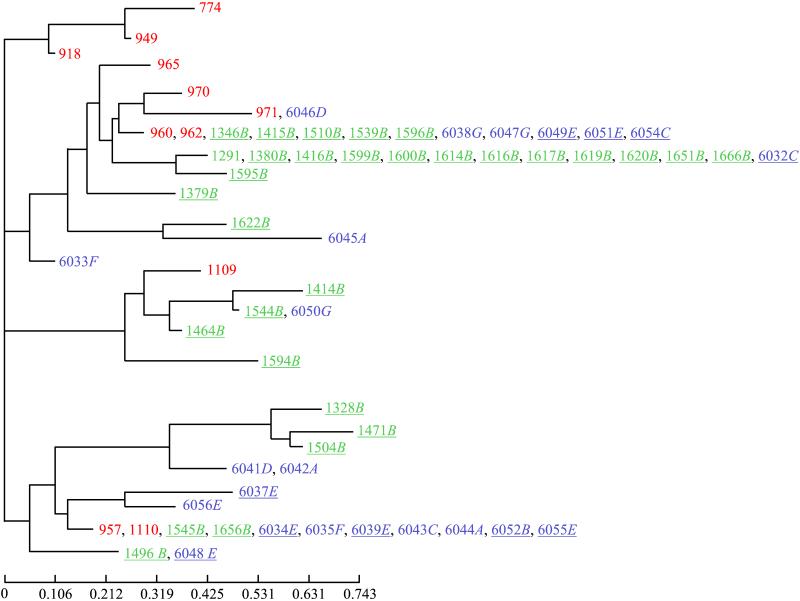
Among the 33 enzymatic loci analyzed, 20 display the same allele for the 64 strains and 13 are polymorphic (Table 2). The unbiased estimate of expected heterozygosity (22) is 0.11. This is a low value compared to that reported for C. albicans (0.17 [29] and 0.35 [1]) and could be attributed to the haploid state of C. glabrata (everything else being equal, haploids display half the genes). There were 26 multilocus genotypes (I to XXVI in Table 2), 7 of which are represented by more than one strain (up to 13, genotype X [Table 2]). Figure 1 gives a representation of the genetic relationships between the different strains according to their geographical origin, anatomic location, and patient pathology. None of the apparent structures in this dendrogram seems to correlate with any of these parameters (Fig. 1 and see below).
TABLE 2.
Genotypes (I to XXVII) observed at the different polymorphic loci analyzeda
| Genotype | Strainsb | No. of loci
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acp | Est-4μ | Fk-1 | Idh | Lap-1 | Lap-2 | Mpi-1 | Pep-B | Pep-C-1 | Pep-D | Pgm-1 | Pgm-2 | Sod-1 | ||
| I | 774 | 4 | 2 | 3 | 1 | 3 | 3 | 4 | 6 | 3 | 4 | 3 | 1 | 1 |
| II | 918 | 4 | 2 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 4 | 3 | 3 | 1 |
| III | 949 | 4 | 2 | 3 | 1 | 3 | 3 | 4 | 6 | 3 | 4 | 3 | 3 | 1 |
| IV | 965 | 3 | 2 | 3 | 3 | 3 | 3 | 4 | 5 | 3 | 3 | 3 | 3 | 1 |
| V | 970 | 3 | 2 | 3 | 3 | 3 | 3 | 4 | 4 | 3 | 1 | 3 | 3 | 1 |
| VI | 1109 | 1 | 1 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 5 | 1 | 3 | 1 |
| VII | 1328 | 4 | 2 | 3 | 3 | 2 | 3 | 3 | 6 | 2 | 2 | 3 | 3 | 1 |
| VIII∗ | 1346 | 3 | 2 | 3 | 3 | 3 | 3 | 4 | 4 | 3 | 3 | 3 | 3 | 1 |
| IX | 1379 | 3 | 2 | 3 | 3 | 3 | 3 | 4 | 1 | 3 | 5 | 3 | 3 | 1 |
| X∗ | 1380 | 3 | 4 | 3 | 3 | 3 | 3 | 4 | 4 | 3 | 3 | 3 | 3 | 1 |
| XI | 1414 | 1 | 1 | 3 | 3 | 2 | 3 | 4 | 6 | 2 | 4 | 1 | 3 | 1 |
| XII | 1464 | 1 | 1 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 4 | 1 | 3 | 1 |
| XIII | 1471 | 4 | 2 | 3 | 3 | 2 | 3 | 3 | 6 | 2 | 3 | 3 | 1 | 1 |
| XIV | 1504 | 4 | 2 | 3 | 3 | 2 | 3 | 3 | 6 | 2 | 3 | 3 | 3 | 1 |
| XV∗ | 1544 | 1 | 1 | 3 | 3 | 2 | 3 | 4 | 6 | 3 | 4 | 1 | 3 | 1 |
| XVI | 1594 | 2 | 1 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 4 | 1 | 2 | 1 |
| XVII | 1595 | 3 | 4 | 3 | 3 | 3 | 3 | 4 | 3 | 3 | 3 | 3 | 3 | 1 |
| XVIII | 1622 | 3 | 2 | 3 | 3 | 3 | 2 | 4 | 6 | 3 | 3 | 3 | 4 | 1 |
| XIX | 6033 | 3 | 2 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 4 | 3 | 3 | 1 |
| XX | 6037 | 1 | 2 | 3 | 3 | 3 | 3 | 3 | 6 | 3 | 3 | 3 | 3 | 3 |
| XXI∗ | 6042 | 1 | 2 | 3 | 3 | 2 | 3 | 4 | 6 | 2 | 3 | 3 | 3 | 1 |
| XXII | 6045 | 3 | 1 | 3 | 3 | 3 | 2 | 4 | 4 | 3 | 4 | 3 | 4 | 1 |
| XXIII∗ | 6046 | 3 | 2 | 2 | 3 | 3 | 3 | 4 | 4 | 3 | 2 | 3 | 3 | 1 |
| XXIV∗ | 6048 | 1 | 2 | 3 | 3 | 3 | 3 | 1 | 6 | 3 | 4 | 3 | 3 | 1 |
| XXV∗ | 6055 | 1 | 2 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 3 | 3 | 3 | 1 |
| XXVI | 6056 | 2 | 2 | 3 | 3 | 3 | 3 | 4 | 6 | 3 | 3 | 3 | 3 | 3 |
Alleles are defined in order of their anodal mobility.
Asterisks indicate repeated genotypes: VIII = strains 960, 962, 1346, 1415, 1510, 1539, 1596, 6038, 6047, 6049, 6051, and 6054; X = strains 1291, 1380, 1416, 1599, 1600, 1614, 1616, 1617, 1619, 1620, 1651, 1666, and 6032; XV = strains 1544 and 6050; XXI = strains 6041 and 6042; XXIII = strains 971 and 6046; XXIV = strains 1496 and 6048; XXV = strains 957, 1110, 1545, 1656, 6034, 6035, 6039, 6043, 6044, 6052, and 6055. Five loci are monomorphic for all of the strains analyzed (i.e., Dia, Gpd, Mpi-2, Pep-A, and Sod-2).
FIG. 1.
Genetic relationships between the different strains of C. glabrata grouped according to (i) their geographical origin (Montpellier in Green, Paris in blue, and reference strains in red), (ii) anatomic location (A, bronchial tubes; B, buccal cavity; C, bronchioalveolar liquid; D, pharynx; E, stools; F, urine; and G, vagina), and (iii) patient pathology (HIV+, underlined). This is an unrooted phenogram built according to the neighbor-joining method applied to the Cavalli-Sforza and Edwards (4) chord distance matrix.
Genetic structure of different samples of C. glabrata.
For this study, reference strains were removed from the analysis. Consequently, Idh (monomorphic in the remaining strains) was not considered in the following.
Nonrandom association between loci.
In Paris, among the 66 possible pairs of loci, six (9%) displayed significant nonrandom association at the 5% level which is not significantly different to what is expected under the null hypothesis (exact binomial test, P = 0.15). Nevertheless, one pair (Acp-PepB, P = 0.000000) remained significant after Bonferroni correction (α′ = 0.05/66 = 0.0008).
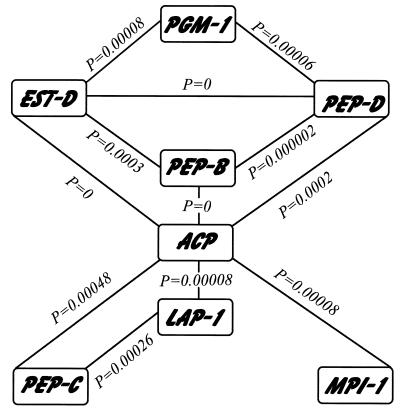
In Montpellier, among the 66 possible pairs of loci, 55 displayed enough polymorphism for a genotypic association test to be done. Among these, 19 pairs (35%) displayed significant nonrandom association at the 5% level, which is far more than expected (P = 0.000, binomial test); 13 of these remained significant after sequential Bonferroni corrections (Fig. 2).
FIG. 2.
The eight loci involved in significantly nonrandom pairs at the Bonferroni level. The exact P value for nonrandom association is given for each pair of loci. Note that Acp and Pgm-1 are not significantly linked together but are strongly linked to each of the remaining loci.
Significance for nonrandom association between multilocus repeated genotypes.
The four most common multilocus genotypes (Paris genotypes VIII and XXV and Montpellier genotypes VIII and X) (Table 2) were analyzed by this test. In each case the probability obtained was highly significant (Table 3). It should be noted that these genotypes are frequently found in more than one place (Paris, Montpellier, or within reference strains). For example, genotype VIII is found in the three groups.
TABLE 3.
Distribution and number of repetitions (n) of multilocus genotypes among Montpellier, Paris, and reference strainsa
| Strain | Paris
|
Montpellier
|
No. of reference strains | ||
|---|---|---|---|---|---|
| n | Probability (d1) | n | Probability (d1) | ||
| VIII | 5 | 8.1 × 10−4 | 4 | 2.3 × 10−2 | 2 |
| X | 1 | - | 12 | 2.8 × 10−8 | 0 |
| XV | 1 | - | 1 | - | 0 |
| XXI | 2 | - | 0 | - | 0 |
| XXIII | 1 | - | 0 | - | 1 |
| XXIV | 1 | - | 1 | - | 0 |
| XXV | 7 | 1.4 × 10−4 | 2 | - | 2 |
| Unique | 4 | 10 | |||
| Overall sample size | 22 | 30 | 5 | ||
The combinatorial probability (d1) of sampling a given multilocus genotype more than or as often as actually observed is provided within each sample for genotypes found at least four times. Unique genotypes were found only once in the whole sample. -, combinatorial probability not calculated.
Multilocus linkage disequilibrium.
The standardized indices of association were 0.0768 and 0.1945 for Paris and Montpellier, respectively. The absence of linkage was strongly rejected in each case (P = 0.0008 and P = 0.0001 for Paris and Montpellier, respectively).
Population structure.
There was a marginally significant differentiation between Paris and Montpellier over all loci (Fst = 0.11, P = 0.054) (Table 4). Three loci (Acp, Est, and Sod [Table 4]) of twelve (25%) displayed a significant Fst that was far more than that expected under the null hypothesis (exact binomial test, P = 0.02). The differentiation between strains from HIV+ and HIV− patients could only be tested in Paris (there was only one HIV− strain in Montpellier; see Table 1). No differentiation was seen at any locus or overall (Fst = 0.015, P = 0.16) (Table 4). Similarly, anatomic location could only be tested in Paris. Here again, no differentiation was suggested from our sample (Fst = 0, P = 0.56) (Table 4). Nevertheless, a statistical linkage could be found between loci. This means that the information carried by these loci is redundant (correlation between loci) and may bias differentiation analysis. Excluding loci that were linked in both samples (i.e., Acp, Pep2, Lap1, and Pep3) did not alter our conclusions. The existence of repeated multilocus genotypes may suggest clonal propagation. It may thus be more appropriate to consider all loci as a whole and each multilocus genotype as the different allelic states of a single locus. Doing so did not considerably change the Fst estimate between Paris and Montpellier (Fst = 0.1), but differentiation became highly significant (P = 0.005). HIV status or anatomic location still appeared to be uninfluential (P = 1.0 and P = 0.7, respectively). It should be noted, however, that our sample sizes could have only detected strong signals. We can thus conclude that no strong differences exist between strains found in different body parts or in different immunological contexts.
TABLE 4.
Fst estimate (θ) and the associated probability required to observe a P value as large as or larger than θ obtained between Paris and Montpellier isolates, between HIV+ and HIV− patients, and between strain samples from the respiratory tract (RT) and from the anal and urogenital spheres (AUG) in Paris
| Locus | Paris (n = 22) and Montpellier (n = 30)
|
HIV+ (n = 11) and HIV− (n = 11)
|
RT (n = 9) and AUG (n = 13)
|
|||
|---|---|---|---|---|---|---|
| θ | P | θ | P | θ | P | |
| Acp | 0.125 | 0.0348 | −0.083 | 0.9999 | −0.094 | 0.9999 |
| Est-4μ | 0.242 | 0.0025 | 0.006 | 0.4756 | −0.028 | 0.6911 |
| Fk | 0.015 | 0.4271 | 0.000 | 0.9999 | 0.043 | 0.4108 |
| Lap-1 | −0.037 | 0.9999 | 0.200 | 0.2115 | −0.009 | 0.5427 |
| Lap-2 | −0.039 | 0.9999 | 0.000 | 0.9999 | 0.043 | 0.4111 |
| Mpi-1 | −0.029 | 0.8184 | 0.050 | 0.474 | 0.006 | 0.9999 |
| Pep-B | 0.066 | 0.1179 | −0.100 | 0.9999 | −0.06 | 0.6617 |
| Pep-C-1 | −0.032 | 0.6935 | 0.100 | 0.4788 | 0.183 | 0.1568 |
| Pep-D | −0.039 | 0.9999 | 0.060 | 0.3258 | −0.064 | 0.4255 |
| Pgm-1 | 0.004 | 0.3753 | 0.000 | 0.9999 | −0.031 | 0.9999 |
| Pgm-2 | −0.022 | 0.9999 | 0.000 | 0.9999 | 0.043 | 0.4111 |
| Sod-1 | 0.269 | 0.0051 | 0.057 | 0.3904 | −0.067 | 0.6773 |
| All loci | 0.089 | 0.0537 | 0.015 | 0.1557 | −0.032 | 0.5621 |
Simulation study.
For the n = 22 case, among the 30 samples obtained, 28 samples (93%) yielded a higher number of significant genotypic association tests (at the 5% or sequential Bonferroni levels) than for C. glabrata in Paris. For the n = 30 samples, the proportion of samples giving higher significant levels than C. glabrata in Montpellier was 16.6% (five samples) for the 5% level of significance and 10% (three samples) for the sequential Bonferroni levels. We conclude that the results obtained for C. glabrata do not significantly differ from those obtained from this simulated purely asexual haploid population. We noticed, however, that the Paris sample displays fewer significant tests compared to the simulation results, in which 20.6% of the tests were significant at the 5% level.
Comparison between the two genetic data sets obtained by MLEE and RAPD.
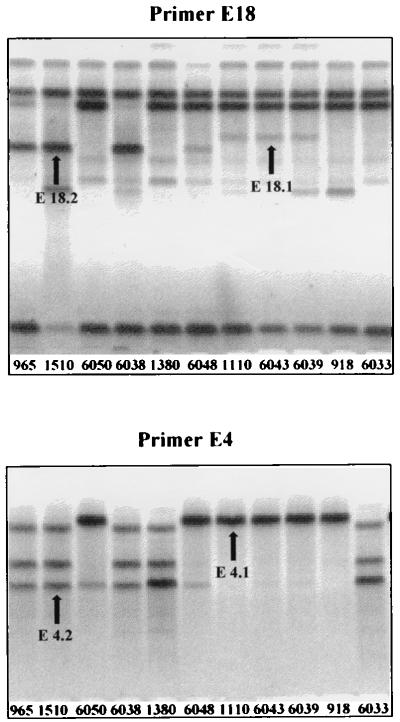
For this study, a subsample of 20 strains (774, 918, 965, 1110, 1328, 1379, 1380, 1414, 1464, 1471, 1510, 6033, 6038, 6039, 6041, 6043, 6045, 6046, 6048, and 6050), corresponding to 17 genotypes, were chosen in order to examine the existing enzymatic variability presented in Fig. 1. For the computation of genetic distances, all 33 loci were taken into account. The RAPD patterns obtained (Fig. 3) are presented in Tables 5 and 6. Geography did not significantly influence genetic distances for enzymes (P = 0.17) or RAPD analysis (P = 0.32) (Mantel tests). This result is in agreement with the low degree of overall loci differentiation previously observed. The correlation between the two matrices obtained for enzymes and RAPD is highly significant (Mantel test, P = 0.00009). This was confirmed by the highly significant correlation between the two half-matrices (RAPD and enzymes) of residuals corrected for geography (Mantel test, P = 0.00006).
FIG. 3.
RAPD patterns (primers E18 and E4) obtained for different strains of C. glabrata (see Materials and Methods). For instance, E18 displays two bands (loci): E18.1, amplified in strain 6043 but not in strain 1510, which amplified band E18.2. Thus, strain 1510 carries allele 0 at locus E18.1 and allele 1 at E18.2.
TABLE 5.
RAPD profiles obtained with the primers of Kit E (see Materials and Methods) for the different strains of C. glabrata tested
| Strain | RAPD profile
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E4.1 | E4.2 | E6.1 | E6.2 | E14 | E17.1 | E17.2 | E17.3 | E18.1 | E18.2 | |
| 774 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| 918 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| 965 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| 1110 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| 1328 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| 1379 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| 1380 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 1414 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 1464 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| 1471 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| 1510 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| 6033 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| 6038 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| 6039 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| 6041 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| 6043 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| 6045 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| 6046 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| 6048 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| 6050 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
TABLE 6.
RAPD profiles obtained with the primers of Kit F (see Materials and Methods) for the different strains of C. glabrata tested
| Strain | RAPD profile
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | F2 | F4.1 | F4.2 | F6 | F10 | F12 | F13 | F14.1 | F14.2 | F14.3 | F16.1 | F16.2 | F16.3 | |
| 774 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 918 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 965 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| 1110 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| 1328 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| 1379 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| 1380 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 |
| 1414 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 1464 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 1471 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| 1510 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| 6033 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 |
| 6038 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| 6039 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| 6041 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 6043 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 |
| 6045 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 |
| 6046 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 |
| 6048 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| 6050 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
In agreement with these results, strains sharing an identical electrophoretic pattern (strains 1110, 6039, and 6043 for genotype XXV and strains 1510 and 6038 for genotype VIII) also appeared to have identical RAPD patterns (Tables 5 and 6).
DISCUSSION
The difference observed between hospitals in Paris and Montpellier reveals that the frequency of multilocus genotypes varies in space and that the “clone flow” is thus restricted to some extent. Nevertheless, the level of differentiation remains low. With larger geographical distances, differentiation would probably have been much more pronounced, as suggested by mitochondrial DNA data for several strains of C. glabrata from Brazil and the United States (36).
For the 63 strains of C. glabrata analyzed, our study reveals the presence of 26 different multilocus genotypes. Among the 33 loci used, no heterozygous profile could be found. Considering the polymorphism observed for these strains, this result supports the haploid chromosomal structure for this yeast, as suggested by other studies (43, 44, 46).
The results obtained with the genetic analysis of C. glabrata allow inferences to be made about the reproductive modality of this yeast. Indeed, (i) the evidence of strong nonrandom association between pairs of loci or overall loci (ii) the existence of multilocus repeated genotypes among different patients and different localities, (iii) the demonstration of a strong correlation between two independent genetic data sets (MLEE and RAPD), and (iv) the confirmation by RAPD of two multilocus repeated genotypes found by MLEE strongly support a mainly clonal mode of reproduction. Such a reproductive strategy thus appears frequent in pathogenic fungi (1, 29, 34; but see also reference 39). This would mean that the multilocus genotype represents the informative and variable character in yeast populations. This mode of reproduction, already recognized for laboratory-reared strains, is confirmed for natural populations by the present study. As confirmed by our simulation study, our data set is fully consistent with what can be expected from a purely clonal population. This, however, cannot exclude episodic sexuality and its consequences on clonal stability through time. A very low rate of recombination could indeed exist. In particular, this may be the case in Paris where statistical association between pairs of loci seemed weaker than expected with pure clonality.
A statistical linkage between loci may be the consequence of a physical tight linkage or to epistatic selection. This interpretation is not supported by our results since our loci are expected to be randomly sampled in the genome. Moreover, the loci involved in statistical linkages are not necessarily the same in Paris and Montpellier; some multilocus genotypes are repeated in different samples (Paris, Montpellier, and reference strains), and a strong correlation exists between allozymes and RAPD markers. These last observations do not support an effect of biased sampling of C. glabrata in humans (Wahlund effect hypothesis) since geography does not seem to be a sufficiently strong structuring factor. Indeed, Fst does not exceed 0.11 between Montpellier and Paris (800 km), and it is not likely that each hospital recruits strains from sufficiently distant areas to promote the required strong Wahlund effect, in particular in Montpellier where many pairs of loci displayed highly significant linkage.
Strong genotypic associations are also expected in the case of the coexistence of numerous and differentiated species in a sample. Although it is difficult to absolutely reject this hypothesis, the lack of any specificity of any clone for any of the available parameters (organs, immunocompetence, etc.) implies that these putative species share identical ecological requirements. In our view, this hypothesis is not the most parsimonious available.
A clonal genetic structure has many consequences for the epidemiology of such pathogens. We did not observe any correlation between particular multilocus genotypes and organ specificity, e.g., the respiratory tract and the anal and urogenitary sphere (Table 4) or associated pathology (HIV+ or HIV−). This means that any clone can be pathogenic in any infection site. In Montpellier, more than one-third of the patients were infected by the same strain. This suggests that nosocomial infection or frequent interpatient transfers may be important factors in the transmission of this yeast. The second point is more likely because many multilocus genotypes are present in distant places. However, we need data from more strains, sampled both within and outside of hospitals, to confirm this.
C. glabrata displays a weak susceptibility to fluconazole (8, 25, 31), leading to many C. glabrata-associated relapses after treatments (46). RFLP typing showed that only specific multilocus genotypes of C. glabrata survive fluconazole treatment (14). Fluconazole treatments thus favor certain clones against the others. Our samples did not allow for a real test, but our data do not suggest any correlation between MLEE genotypes and fluconazole resistance (Table 1 and Fig. 1). The mutation rate of resistance may be high (appearing in association with many MLEE combinations), or antifungal treatments may be insufficient in the hospitals involved in this study. Sexual reproduction involving resistant clones may prevent resistant genes from being “marked” by other loci. However, further investigations are needed before definite conclusions can be drawn.
Acknowledgments
We thank Patrick Durand for help in genetic distance computations for RAPD markers, Austin Burt for very useful comments on an early version of the manuscript, Phil Agnew for critical reading of the manuscript, and two anonymous referees who helped considerably to improve the manuscript.
REFERENCES
- 1.Boerlin, P., F. Boerlin-Petzold, J. Goudet, C. Durussel, J. L. Pagani, J. P. Chave, and J. Bille. 1996. Typing Candida albicans oral isolates from human immunodeficiency virus-infected patients by multilocus enzyme electrophoresis and DNA fingerprinting. J. Clin. Microbiol. 34:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borg-von Zepelin, M., H. Eiffert, M. Kann, and R. Rüchel. 1993. Changes in the spectrum of fungal isolates: results from clinical specimens gathered in 1987/88 compared with those in 1991/92 in the University Hospital Göttingen, Germany. Mycoses 36:247-253. [DOI] [PubMed] [Google Scholar]
- 3.Caugant, D. A., and P. Sandven. 1993. Epidemiological analysis of Candida albicans strains by multilocus enzyme electrophoresis. J. Clin. Microbiol. 31:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalli-Sforza, L. L., and A. W. F. Edwards. 1967. Phylogenetic analysis: model and estimation procedures. Am. J. Hum. Genet. 19:233-257. [PMC free article] [PubMed] [Google Scholar]
- 5.Dodd, C. L., D. Greenspan, M. H. Katz, J. L. Westenhouse, D. W. Feigal, and J. S. Greenspan. 1991. Oral candidiasis in HIV infection: pseudomembranous and erythematous candidiasis show similar rates of progression to AIDS. AIDS 5:1339-1343. [PubMed] [Google Scholar]
- 6.Dupont, B., J. R. Graybill, D. Armstrong, R. Laroche, J. E. Touzé, and L. J. Wheat. 1992. Fungal infections in AIDS patients. J. Med. Vet. Mycol. 30:19-28. [DOI] [PubMed] [Google Scholar]
- 7.Fan-Havard, P., D. Capano, S. M. Smith, A. Mangia, and R. H. Eng. 1991. Development of resistance in Candida isolates from patients receiving prolonged antifungal therapy. Antimicrob. Agents Chemother. 35:2302-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, R., K. R. Neal, C. L. S. Leen, M. E. Ellis, and B. K. Mandal. 1991. Fluconazole resistant Candida in AIDS. J. Infect. 22:201-204. [DOI] [PubMed] [Google Scholar]
- 9.Goudet, J. 1995. Fstat version 1.2: a computer program to calculate F-statistics. J. Hered. 86:485-486. [Google Scholar]
- 10.Goudet, J., T. de Meeus, A. J. Day, and C. J. Gliddon. 1994. The different levels of population structuring of the dogwhelk, Nucella lapillus, along the South Devon Coast, p. 81-95. In A. R. Beaumont (ed.),Genetics and evolution of aquatic organisms. Chapman & Hall, London, England.
- 11.Goudet, J., M. Raymond, T. de Meeûs, and F. Rousset. 1996. Testing differentiation in diploid populations. Genetics 144:1933-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-849. [DOI] [PubMed] [Google Scholar]
- 13.Haubold, B., M. Travisano, P. B. Rainey, and R. R. Hudson. 1998. Detecting linkage disequilibrium in bacterial populations. Genetics 150:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitchcock, C. A., G. W. Pye, P. F. Troke, E. M. Johnson, and D. W. Warnock. 1993. Fluconazole resistance in Candida glabrata. Antimicrob. Agents Chemother. 37:1962-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitchen, V. S., M. Savage, and J. R. W. Harris. 1991. Candida albicans resistances in AIDS. J. Infect. 22:204-205. [DOI] [PubMed] [Google Scholar]
- 16.Klein, R. S., C. A. Harris, C. Small, B. Moll, M. Lesser, and G. H. Friedland. 1984. Oral Candidiasis in high risk patients as the initial manifestation of the acquired immunodeficiency syndrome. New Engl. J. Med. 311:354-358. [DOI] [PubMed] [Google Scholar]
- 17.Korting, H. C., M. Ollert, A. Geogii, and M. Froschl. 1988. In vitro susceptibilities and biotypes of Candida albicans isolates from the oral cavities of patients infected with human immunodeficiency virus. J. Clin. Microbiol. 26:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacFarlane, T. W. 1990. Ecology and epidemiology of Candida, p. 21-46 . In L. P. Samaranayake and T. W. MacFarlane (ed.), Oral candidosis, Wright, London, England.
- 19.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Manly, B. F. J. 1985. The statistics of natural selection, p. 176-180. Chapman & Hall, New York, N.Y.
- 21.Nei, M. 1972. Genetic distance between populations. Am. Nat. 106:283-291. [Google Scholar]
- 22.Nei, M. 1978. Estimate of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei, M., and J. C. Miller. 1990. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 125:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, T. T. C., and D. W. Denning. 1993. Fluconazole resistance in Candida in patients with AIDS: a therapeutic approach. J. Infect. 26:117-125. [DOI] [PubMed] [Google Scholar]
- 26.Odds, F. C. 1988. Candida and candidiosis,2nd ed., p. 279-313. Leicester University Press, Leicester, England.
- 27.Pasteur, N., G. Pasteur, F. Bonhomme, J. Catalan, and J. Britton-Davidian. 1987. Manuel technique de génétique par electrophorèse des protéïnes. Lavoisier, Paris, France.
- 28.Pujol, C., J. Reynes, F. Renaud, M. Mallié, and J. M. Bastide. 1993. Analyse génétique de souches de Candida albicans par électrophorèse des isoenzymes. J. Mycol. Med. 3:14-19. [Google Scholar]
- 29.Pujol, C., J. Reyne, F. Renaud, M. Raymond, M. Tibayrenc, F. J. Ayala, F. Janbon, M. Mallié, and J. M. Bastide. 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90:9456-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymond, M., and F. Rousset. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86:248-249. [Google Scholar]
- 31.Regli, P., A. Blancard, M. Goudard, J. Moulin-Traffort, J. M. Sarzier, and M. Quilici. 1992. Evaluation de la sensibilité in vitro au fluconazole de différentes espèces de levures rencontrées en pathologie. Pathol. Biol. 40:500-506. [PubMed] [Google Scholar]
- 32.Rice, W. R. 1989. Analysing tables of statistical tests. Evolution 43:223-225. [DOI] [PubMed] [Google Scholar]
- 33.Richardson, B. J., P. R. Baverstock, and M. Adams. 1986. Allozyme electrophoresis: a handbook for animal systematics and population studies. Academic Press, Sydney, Australia.
- 34.Rodriguez, E., T. de Meeûs, M. Maillé, F. Renaud, F. Symoëns, P. Mondon, M. A. Piens, B. Lebeau, M. A. Viviani, R. Grillot, N. Nolard, F. Chapuis, A. M. Tortorano, and J. M. Bastide. 1996. Multicentric epidemiological study of Aspergillus fumigatus isolates by multilocus enzyme electrophoresis. J. Clin. Microbiol. 34:2559-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Sanson, G. F. O., and M. R. S. Briones. 2000. Typing of Candida glabrata in clinical isolates by comparative sequence analysis of the cytochrome c oxydase subunit 2 gene distinguishes two clusters of strains associated with geographical sequence polymorphisms. J. Clin. Microbiol. 38:227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slatkin, M. 1987. Gene flow and the geographic structure of natural populations. Science 236:787-792. [DOI] [PubMed] [Google Scholar]
- 38.Takezaki, N., and M. Nei. 1996. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, J. W., D. M. Geiser, A. Burt, and V. Koupopanou. 1999. The evolutionary biology and population genetics underlying fungal strain typing. Clin. Microbiol. Rev. 12:126-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tibayrenc, M. 1998. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int. J. Parasitol. 28:85-104. [DOI] [PubMed] [Google Scholar]
- 41.Tibayrenc, M., F. Kjellberg, and F. J. Ayala. 1990. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. USA 87:2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weir, B. S., and C. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358-1370. [DOI] [PubMed] [Google Scholar]
- 43.Whelan, W. L., and K. J. Kwon-Chung. 1987. Parasexual genetics of Torulopsis glabrata. J. Bacteriol. 169:4991-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whelan, W. L., R. M. Partridge, and P. T. Magee. 1980. Heterozygosity and segregation in Torulopsis albicans. Mol. Gen. Genet. 180:107-113. [DOI] [PubMed] [Google Scholar]
- 45.Whelan, W. L., S. Simon, E. S. Beneke, and A. L. Rogers. 1984. Auxotrophic variants of Torulopsis glabrata. FEMS Microbiol. Lett. 24:1-4. [Google Scholar]
- 46.Wingard, J. R., W. G. Merz, M. G. Rinaldi, C. B. Miller, J. E. Karp, and R. Saral. 1993. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob. Agents Chemother. 37:1847-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright, S. 1931. Evolution in Mendelian populations. Genetics 16:97-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, J., T. G. Mitchell, and R. Vigalys. 1999. PCR-restriction fragment length polymorphism (RFLP) analyses reveal both extensive clonality and local genetic differences in Candida albicans. Mol. Ecol. 8:59-73. [DOI] [PubMed] [Google Scholar]