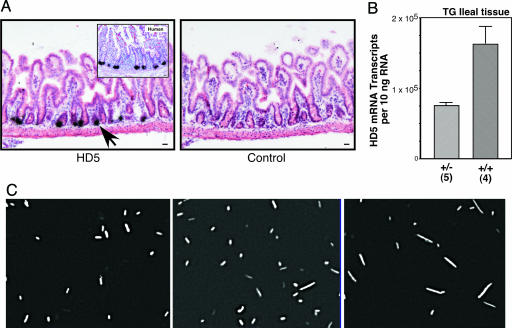
Fig. 4.
Bacterial microbiota in wild-type, heterozygous, and homozygous HD5 TG mice. (A) Expression and localization of HD5 mRNA in TG mouse small intestine analyzed by in situ hybridization by using an antisense probe (Left). Hybridization of HD5 probe to section of human ileum (Inset). Wild-type mouse shows no hybridization to HD5 antisense probe (data not shown). Sense probe (Right) and RNase A pretreatment controls (data not shown) were negative for hybridization signal. Arrows point to dense signals that overlie PCs. Counterstain was hematoxylin/eosin. (Scale bar, 20 μm.) (B) Expression of HD5 mRNA in ileal specimens from heterozygous and homozygous HD5 TG mice. Data expressed as mRNA copy number per 10 ng of total RNA determined with quantitative real-time RT-PCR using external standards. [Scale bars represent means (±standard error).] (C) FISH analysis of luminal microbes in mouse ileum. Representative hybridization analysis with TR-Bac338 probe (detecting all bacteria) is shown for wild-type mice (Left), HD5 TG heterozygote mice (Center) and HD5 TG homozygote mice (Right).