Abstract
A living cell is not an aggregate of molecules but an organized pattern, structured in space and in time. This article addresses some conceptual issues in the genesis of spatial architecture, including how molecules find their proper location in cell space, the origins of supramolecular order, the role of the genes, cell morphology, the continuity of cells, and the inheritance of order. The discussion is framed around a hierarchy of physiological processes that bridge the gap between nanometer-sized molecules and cells three to six orders of magnitude larger. Stepping stones include molecular self-organization, directional physiology, spatial markers, gradients, fields, and physical forces. The knowledge at hand leads to an unconventional interpretation of biological order. I have come to think of cells as self-organized systems composed of genetically specified elements plus heritable structures. The smallest self that can be fairly said to organize itself is the whole cell. If structure, form, and function are ever to be computed from data at a lower level, the starting point will be not the genome, but a spatially organized system of molecules. This conclusion invites us to reconsider our understanding of what genes do, what organisms are, and how living systems could have arisen on the early Earth.
“All this has been said before—but since nobody listened, it must be said again.” — André Gide
INTRODUCTION
Spatial organization is perhaps the most conspicuous quality of cells and organisms, and also the most elusive. The dissection of life into its molecular constituents—the genes, enzymes, and lipid bilayers—necessarily starts with the destruction of spatial order. Yet we know that most complex functions, such as motility and division, depend on having the right molecules in their proper places. Architecture is what ultimately distinguishes a living cell from a soup of the chemicals of which it is composed. How cells generate, maintain, and reproduce their spatial organization is central to any understanding of the living state.
We have, of course, a formal framework for thinking about such matters: organization, like other aspects of physiology and function, manifests the instructions encoded in the genes. This view of life, given classical expression by François Jacob (75), pervades both the technical and popular literature. Genes, acting individually or through elaborate regulatory networks (3, 110), specify the structure of living matter and ensure its persistence; ultimately it is the genes that build cells. At the chemical, compositional level, genes do, indeed, specify much of cellular organization. The question that concerns us here is how living things bridge the gap between the molecules on the nanometer scale and cells on the scale of micrometers to millimeters.
Over the past 15 years, spatial organization has moved to the forefront of research in cell biology; no longer would it be fair to say, as I did in 1990 (57), that “of cellular morphogenesis … we know much but understand little.” And yet, it remains as difficult as ever to extract enlightenment from the torrent of data. Is cellular architecture explicitly spelled out by genes, and if so, how? If not, how is spatial organization passed from one generation to the next? How do molecules find the correct location in the cell space? What is the origin of large-scale order, as illustrated by the mitotic spindle or the endomembrane system of eukaryotic cells? How do multitudes of molecules reproducibly come together into cellular forms, which in turn serve as the targets of natural selection? And is it true that, at least in principle, nothing irretrievable would be lost if a cell were carefully dissociated into its molecular constituents? These questions define the scope of the present article.
Data seldom speak for themselves: they are apt to be unintelligible in the absence of a conceptual framework to put the information in order. I have therefore tried to distill from the literature a set of broad and comprehensible principles to explain how molecules come together into cellular systems that are spatially organized, functionally coherent, and competitive in the evolutionary arena and to illustrate these principles by examples drawn from recent research with both prokaryotic and eukaryotic microorganisms. This exercise provides fresh support for a holistic point of view that diverges significantly from the opinions held, at least conventionally, by many molecular scientists. Spatial organization is not written out in the genetic blueprint; it emerges epigenetically from the interplay of genetically specified molecules, by way of a hierarchy of self-organizing processes, constrained by heritable structures, membranes in particular. Molecules and the genes that specify them remain essential since they constitute the material basis of all biological structures, but from the perspective of organized systems they do not hog the limelight. Center stage is held by the whole cell, that indispensable unit of life, of which molecules are but parts; the smallest self that truly organizes itself is the cell.
Nothing that can be said in science is without precedent. In addition to the technical papers cited below, I have drawn freely upon the ideas of many colleagues (2, 22, 23, 42, 48, 51, 52, 86, 121, 151). Many of the present musings have surfaced in my own earlier writings, too (57, 58, 60, 61, 62); but the case deserves to be made anew, for the basis upon which it rests grows ever richer and more solid.
THE HIERARCHY OF ORDER
Beyond the Genes
In scientific parlance the terms heredity and genetics have become practically synonymous: features that are reproduced from one generation to the next are said to be hereditary and assumed to be encoded in gene sequences. At the molecular level, the level of proteins and nucleic acids, few will quarrel with this sweeping generalization except to make room for complexities, but it is the organization of the cell as a whole that brings the point into focus. Cell form and cytology are obviously inherited across thousands of generations. How much of the architecture of prokaryotic and eukaryotic cells will be explicitly spelled out in their genes?
To make the issue more concrete, consider what cell growth and division entail. The first requirement is to duplicate all of the cell's molecular constituents. These structures are specified by the genes, directly or indirectly, together with much regulatory machinery (note, though, that even here there is room for ambiguity: only by implication is the chemical structure of peptidoglycan or of lipopolysaccharide written out in the genes that encode the enzymes which produce those molecules). The genetic instructions often include information pertinent to the localization of the product. Targeting sequences direct proteins to the plasma membrane, nucleus, mitochondria, or lysosomes. Certain proteins and mRNAs are transported individually to particular locations in cell space, and this localization depends on having an appropriate sequence. Transport vesicles recognize specific target membranes, such as the Golgi, vacuole, or plasma membrane, with the aid of SNARE proteins. But there is much more to growth and division than manufacturing the parts. A rod-shaped cell must also elongate with constant diameter, construct an efficient apparatus to partition its chromosomes, locate its midpoint, lay down a septum, and undergo fission. In eukaryotic cells, targeted vesicle fusion requires, in addition to the SNAREs, both a delivery system and a secretory apparatus. Is all this elaborate choreography spelled out in particular genes? Evidently not, for the many genomes now on record apparently contain no genes that specify cellular forms and patterns. Genes specify the molecular parts, not their arrangement into a higher order.
In speaking of these matters, I have sometimes encountered the objection that many mutations are known to alter the form and spatial organization of cells. Does that not demonstrate that cellular architecture comes under the genes' writ? Well, yes and also no. If a protein deleted or altered by mutation plays a role in morphogenesis, the mutant's form or organization may well be affected. Examples from both prokaryotes and eukaryotes run into the hundreds (39, 40, 57, 58, 64, 65, 100, 131, 135). Such mutants are immensely valuable in dissecting morphogenetic pathways, but they do not show that cell form and organization are explicitly spelled out in the genetic instructions. Indeed, I argue as a matter of principle that cell morphology cannot be read out of genomic sequences alone.
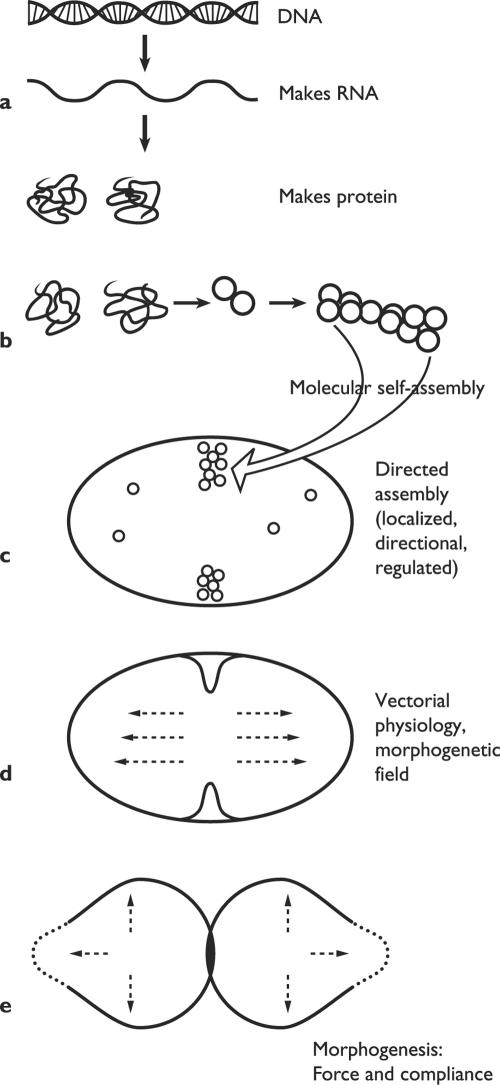
A more realistic framework for reflection on the genesis of biological organization and morphology is sketched in Fig. 1. The hierarchy of order envisages a nested succession of stages, beginning with the translation of genetic information into functional proteins. Various kinds of self-assembly give rise to subcellular structures and devices. Next come the localized and vectorial processes of physiology, all subordinated to the structure of the cell as a whole, which generate spatial patterns on a scale orders of magnitude above the molecular. The hierarchy culminates with the generation and application of the mechanical forces that actually shape the whole cell or microorganism. Organisms are notoriously diverse, and they have invented a host of ways to shape themselves. If unity can be discerned, it revolves around the kinds of processes that progressively build up structures, organization, and global form. The word to conjure with nowadays is self-organization.
FIG. 1.
Hierarchy of biological order. The spatial and functional organization of cells is produced by a nested succession of processes that bridge the gap between the molecular scale and the cellular one. (a) DNA sequences are transcribed into RNA and then translated into amino acid chains; the latter fold spontaneously into functional proteins. (b) Ribosomes and other supramolecular complexes arise by self-assembly of their molecular constituents. (c) Cellular structures arise in a controlled manner at a particular time and place. (d) Many physiological processes have a direction in cell space. (e) Morphogenesis results from local compliance with applied forces, such as turgor pressure. (Modified from Harold [61].)
Self-Organization
Travelers in alpine and arctic regions sometimes pass through the puzzling landscapes known as “patterned ground”: numberless stones neatly arranged in rows, circles, or polygons that may extend for miles. It is not human hands that have put these stones in order, but physical forces alone. Kessler and Werner (83) have developed numerical models that account for this landform by the interplay of two forces: pressure generated by freezing and thawing of the soil, coupled with redistribution of the upheaved stones by gravity. The spontaneous emergence of macroscopic patterns, in the absence of directive or design, contradicts our well-founded prejudice that, when left to themselves, things are more likely to fall apart than to put themselves in order. Self-organization is nevertheless widespread in nature and is observed on all scales (7). We have galaxies and hurricanes, crystals and chemical waves, and a growing roster of synthetic objects and materials shaped by self-assembly (95, 161). Biologists emphasize the role of self-organization in the behavior of schools of fish and flocks of birds and in the way termites construct those astonishing mud towers on the jungle floor (21). And one can make a strong case for assigning self-organization a major role in generating spatial order on the cellular scale (113).
For the purposes of cell biology, let me define self-organization as the emergence of supramolecular order from the interactions among numerous molecules that obey only local rules, without reference to an external template or global plan. This definition is modified from that offered by Camazine (21) and differs significantly from Misteli's (113) in omitting any reference to function. The definition explicitly excludes order imposed by an external template, whether physical (as in a photocopier) or genetic (as in the specification of an amino acid sequence by a sequence of nucleotides).
Examples of self-organization have long been familiar to biochemists under the heading of self-assembly. Ribosomes, microtubules, microfilaments, virus particles, and lipid bilayers come to mind; even the folding of a nascent polypeptide chain into its three-dimensional form can be put into this category. The hallmark of self-assembly is that, at least in principle, it requires no input of either information or energy: self-assembly proceeds down the thermodynamic hill towards equilibrium, or at least towards a free-energy minimum. In practice, there is often room to quibble over the details (would phosphorylation of a protein monomer represent energy input?), but the principle is useful. The structure of the self-assembled complex is wholly specified by the structures of its parts and is therefore implicit in the genes that specify those parts: natural selection crafted those genes to specify parts that assemble into a functional complex.
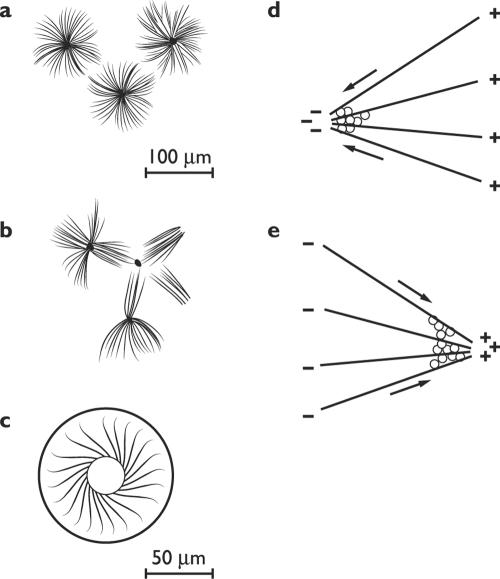
A more versatile category of self-organizing processes may be called dynamic self-assembly (162), or self-construction. When Eric Karsenti and his colleagues add together microtubules, ATP, GTP, and the motor protein kinesin, the components assemble into structures reminiscent of the asters of the mitotic spindle (Fig. 2). With kinesin, a motor protein that moves towards the plus end of microtubules, the minus ends face out; when the minus-directed motor protein dynein is used, the plus ends face out. These structures arise when multimeric molecules of the motor protein bridge two microtubules and move along them, with concurrent hydrolysis of ATP (71). The configurations produced depend on the combination of motor proteins and their concentration, and include vortices and networks (Fig. 2) (118, 152). Efforts to reconstitute spindles continue (81); no one has yet produced an entire spindle from purified components, but since spindles do assemble themselves around injected DNA in extracts made from frogs' eggs, eventual success is almost ensured.
FIG. 2.
Self-organized microtubule patterns. Left, forms. (a) Multimeric kinesin only. (Adapted from Surrey et al. [152] with permission of the publisher.) (b) Multimeric kinesin plus a multimeric Drosophila motor protein. (Adapted from Surrey et al. [152] with permission of the publisher.) (c) Modified kinesin in a toroidal chamber. (Adapted from Nèdèlec et al. [118] with permission of the publisher.) Right, mechanisms. (d) A minus-directed motor (dynein) points the plus ends outward. (Adapted from Karsenti and Vernos [81] with permission of the publisher.) (e) A plus-directed motor (kinesin) points the minus ends outward. (Adapted from Karsenti and Vernos [81] with permission of the publisher.)
Self-construction differs from self-assembly in that its products are not static; their stability reflects a dynamic steady state, the continuous flux of parts in and out of the structure. External directions are not required, but energy must be consumed all the time. Insofar as the form of that steady state depends on that of its constituent molecules (e.g., the polarity of tubulin and of the motor proteins), we are still in the realm governed by the genes. But a cluster of microtubules resembling an aster must be a very remote implication of the primary amino acid sequence of tubulins and would never have been predicted from that sequence.
Self-constructed dynamic patterns are apparently very common and probably underlie much of intracellular organization. Not only the mitotic spindle, but the entire cytoskeleton of eukaryotic cells may be of this nature, including microfilament meshworks as well as microtubules. The same holds for the endomembrane system, the Golgi apparatus in particular, whose assembly from precursor vesicles can be recapitulated in vitro (35, 70, 85, 86, 113). Among prokaryotes, the manner in which rod-shaped cells locate their midpoint and put the divisome in place comes under this heading (see below).
Self-organization, in the sense of the definition proposed at the beginning of this section, is popular because it can be recreated in the test tube and meshes smoothly with the reductionist paradigm. But in fact, few if any of the complex internal structures can arise solely in obedience to local rules, without reference to the function of the whole cell. Spindles, we know, form at particular locations, and cells go to great lengths to ensure their correct orientation; accurate division hinges on it. Centrosomes, which nucleate the microtubules that organize themselves into a cytoskeleton, do so by providing some kind of template. Self-construction is clearly part of Golgi duplication, but there is evidence that it takes place on a matrix or template that carries over from one generation to the next (145); and there are organisms whose Golgi apparatus undergoes what looks remarkably like true division (77, 115). Biological self-organization is real and important, but when it takes place in a living cell it is subject to constraint and control by the system as a whole. The only self that can truly be said to organize itself is the cell.
Directions in Space
Chemical reactions in the living cell, unlike those in the test tube, commonly have both location and orientation in space. Vectorial transport of electrons and protons across membranes, which lies at the heart of energy transduction (Fig. 3a), was an early example. The machinery of DNA replication, transcription, and translation has a direction with respect to the polynucleotide chains, which was made visible in classic electron micrographs. The direction of biochemical processes ultimately derives from the asymmetric structure of proteins and other macromolecules, which then assemble into polarized complexes, including chromosomes, microfilaments, microtubules, and the motor proteins that travel those tracks. To the extent that directionality is rooted in molecular structure, we are still in the orbit of the genes that encode those molecules.
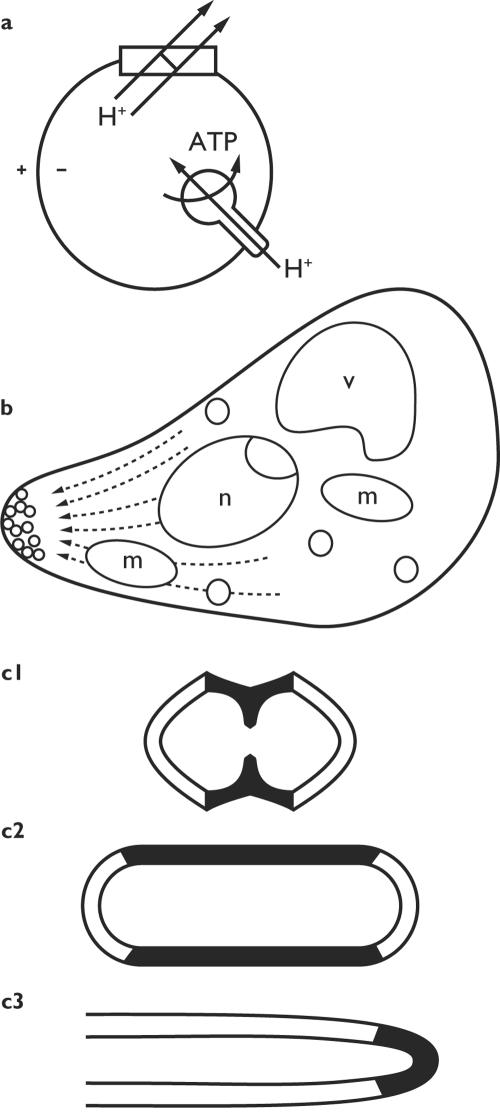
FIG. 3.
Direction and location. (a) Vectorial metabolism. Protons are pumped outward by the respiratory chain and return via the ATP synthase. (b) A polarized cell, with much of its physiology directed towards the transport of secretory vesicles to the tip. n, nucleus; v, vacuole; m, mitochondrion. Based on an electron micrograph of a shmoo of S. cerevisiae by Baba et al. (6). (c) Three patterns of localized wall deposition in growing bacteria: zonal, dispersed, and apical.
Vectorial physiology, directionality at the systems level (57), comes to the fore when one considers the spatial organization of the cell as a whole. Few cells are spherically symmetrical, and even those must break symmetry in order to divide. Growing cells commonly have a durable axis, and most are overtly polarized; Caulobacter crescentus supplies a striking eubacterial example and will be discussed in more detail later. Polarity goes well beyond the visible differences of form and function between one end of the cell and the other. Rather, it implies that ultrastructure and physiology are so arranged as to confer global direction upon all cellular operations (Fig. 3b). Directions in cell space are rooted in the asymmetry of molecules but are established at a higher level of organization, visibly so in cases where physiological vectors arise de novo. The budding yeast Saccharomyces cerevisiae and the germination of the zygotes of brown algae (now known as Fucus and Silvetia spp.) make familiar examples to be discussed below. In eukaryotes, at least, cell polarization normally (perhaps always) turns on the construction of a polarized cytoskeleton.
Developmental mechanisms generally proceed not from the nucleus outward, but from the periphery inward. Lionel Jaffe emphasized this point almost four decades ago (76), and it proved to be a very general principle. The direction is usually supplied by cues from the environment, such as a gradient of pheromone or the direction of incident light; but it may also represent the amplification of a random fluctuation or the execution of a default pathway. In all cases, polarity begins with the establishment of a landmark at the cell surface (34, 57, 120). This serves as the focus upon which the cytoskeleton becomes oriented and to which cytoplasmic transport and/or mechanical forces are directed. A common outcome is the establishment of a localized and directional secretory pathway, the essential physical basis of polarized growth in eukaryotic cells. Whether this generalization also explains the different modes of localized growth in bacteria (Fig. 3c) remains to be seen.
All Connected Together
Take one of the larger ciliates, slice it in two with a sharp blade, and prepare to be surprised. Rather than suffering swift death as its innards spill out, the cell rounds up, reseals its surface, and may even in time regenerate a smaller version of its original form. The capacity of many cells to survive grievous injury has been known for well over a century but remains unappreciated, perhaps because it is so sharply at variance with the classical notion of a fluid cytoplasm. What holds the cell contents together, apparently including ions and small molecules, is not altogether understood (see references 98 and 111 for mainstream reviews and 127 for a radical counterblast), but some things are clear. First, the consistency of eukaryotic cytoplasm is that of a gel whose protein concentration approaches the limit of solubility. And second, an extensive network of cables, fibers, and struts pervades the cytoplasm, linking cell envelope and organelles into a fully connected web. Coherence is one of the gifts of the cytoskeleton.
Microtubules and microfilaments of eukaryotic cells are most conspicuous when serving as the structural foundation of organelles such as cilia, filopodia, and the feeding baskets of ciliates. But the more general role of the cytoskeleton is to integrate cell space, by providing defined pathways for the transmission of materials, forces, and communications. Both microtubules and microfilaments serve as tracks for the transport of organelles, vesicles, other filaments, and individual macromolecules. They also underpin mechanical coupling between distant elements, and they reach across the plasma membrane to link up with the cell wall or the external matrix. In animal cells, and perhaps others also, a web of delicate fibers made of actin penetrates every cranny. The functions of the filamentous structures overlap but are not the same: the microfilaments are thought to resist tension, while microtubules (especially when bundled) resist compression and serve as struts. The cytoskeletal scaffold is flexible, but taut rather than flabby; its stability is dynamic and commonly depends on continuous energy dissipation.
What about prokaryotic cells? The classical cartoon, of a homogenous and fluid cytoplasm in which molecules large and small are free to diffuse at random, is being transformed as powerful new imaging technology reveals more and more structure. The information at hand comes chiefly from eubacteria; archaeal cytoplasm is no less structured, but the molecular specifics may prove to be significantly different. Most bacterial proteins may be mobile, but a growing number are associated with particular locations, at least transiently; histidine kinases are among the proteins whose function depends on being in the right place at the right time (49, 101, 146, 147). And eubacteria possess a cytoskeleton after all. First came the Z-ring (or divisome), a transient cytoskeleton involved in cytokinesis (40, 100, 105, 117); its chief protein constituent, FtsZ, is homologous to tubulin. This was quickly followed by the discovery of helical filaments lying just beneath the plasma membrane; their chief proteins, MreB and Mb1, are homologous to actin (39, 40, 49, 50, 78). And now it appears that even intermediate filaments have eubacterial homologs (5). We do not yet know just what those filaments do, but they are strongly implicated in cell morphogenesis. As to the archaea, homologs of FtsZ and MreB are present in some but not in all; comparative studies of the cytoskeleton promise new insight into the origin of eukaryotic cells.
In a recent historical memoir, Schliwa (144) recounts how every technical advance in the study of the cytoplasm has revealed further structural complexity. Now we are coming to appreciate the web of linkages that create a coherent, functional ensemble out of apparently independent elements. Nothing illustrates this better than a series of articles by Ingber (73, 74), who, over the past two decades, has fleshed out a mechanical model of animal cells based on tensegrity architecture. This approach has made intelligible many features of cell shape and motility, and it now appears that the mechanics of the cytoskeleton impinge on metabolism, signal transduction, and even the execution of genetic programs.
The Continuity of the Cell
The foundations of cell biology, a discipline distinct from its molecular cousin in both subject matter and attitude, were laid in 1858 by Rudolf Virchow, with the proclamation that every cell comes from a previous cell: Omnis cellula e cellula. There is more to this venerable phrase than the recognition that it takes a cell to make a cell; Virchow's law implies that every cell is structurally, architecturally, continuous with its parent cell. For the past three billion years, at least, no cell has ever arisen by direct assembly from its molecular constituents. If Craig Venter and Hamilton Smith succeed in their effort to produce a synthetic cell capable of self-reproduction and evolution (107), they will have created something new under the sun! In the meanwhile, cells will continue to grow and divide in the time-honored manner, building themselves upon the structural framework supplied by an existing cell. The object of this section is to consider just what, apart from genes and gene products, is transmitted from one generation to the next.
Heritable membranes.
Next to the genes, the most important legacy that cells pass on to their offspring are the membranes that make up so much of cytoplasmic architecture. It is a most curious fact, known from the early days of electron microscopy but seldom mentioned in the literature: phospholipid bilayer membranes readily self-assemble in the test tube, but rarely if ever do so in the living cell. On the contrary, the major classes of cellular membranes (plasma membrane, endoplasmic reticulum, nuclear membrane, and those of mitochondria and chloroplasts) all grow, and they grow by extension of an existing membrane. Polarity and membrane type are maintained during growth. Cavalier-Smith, who has made much of membrane heredity in recent years (22, 23), distinguishes between “genetic” membranes, which always arise by growth and division of membranes of the same type (e.g., the plasma membranes of bacteria and the inner and outer mitochondrial membranes), and “derived” ones, which form by differentiation from dissimilar membranes (e.g., the eukaryotic plasma membrane). Genetic membranes, like DNA, appear to have been passed from one generation to the next since the dawn of cellular life.
How membranes grow is generally well understood. In the case of eubacteria, membrane phospholipids are produced by biosynthetic enzymes embedded in that membrane and incorporated in situ. Eukaryotic cells generate phospholipids in the endoplasmic reticulum and initially incorporate them into those membranes. Membrane vesicles then bud off the endoplasmic reticulum, are processed in the Golgi apparatus and redistributed to other destinations such as the plasma membrane or a vacuole. Secretory vesicles find their target with the help of specific receptor proteins, such as the v- and t-SNARES. These membrane proteins, and biosynthetic proteins too, must be targeted to their cognate membranes, recognizing ligand proteins or specific lipids. It is this network of mutual recognition that maintains the topology and identity of each membrane type and ensures that every membrane comes from a previous membrane.
Biology is so riddled with exceptions that a sweeping proclamation of the conservation of membranes naturally invites skepticism. Indeed, potential exceptions do crop up in the literature. The origin of yeast autophagosomes is not fully understood and may just possibly represent an instance of de novo membrane formation (122). One must also wonder about the reconstitution of plasma membranes by cytoplasmic fragments, recently reported for certain giant marine algae (84). There may be other examples that I have failed to find. It seems, however, that these are exceptions that probe the rule without overturning it: the general rule is that membranes grow by enlargement of an existing membrane.
What is true of membranes commonly applies to cell envelopes and walls, perhaps to the cell cortex, though probably not with such rigor. Take, for example, the peptidoglycan walls of bacteria. Cells stripped of their wall can often grow in high-osmolarity medium; L-forms, as they are called, are spherical, devoid of any visible cell wall, and resistant to antibiotics that inhibit wall biosynthesis (123). Some strains can revert to the normal, walled state and bacillary form, and it has been proposed that the new wall must be laid down upon a foundation of peptidoglycan seed molecules. Indeed, Höltje (69) makes a case for considering the peptidoglycan sacculus as a template that directs the biosynthesis of new wall and thus determines the shape of the growing cell. One can make a parallel argument for the outer, lipopolysaccharide membrane of gram-negative bacteria. And in the gram-positive bacterium Enterococcus hirae, the prominent wall band around the circumference visibly undergoes duplication at the time of division (cited in reference 57). Hereditary propagation of such features may not be an absolute requirement, but it does appear to be the norm.
Structural inheritance.
The continuity of membranes and other envelope elements underlies several intriguing genetic tricks, including inheritance independent of nucleic acids and the transmission of acquired characteristics. The star performers in this arena are the ciliates; the field was pioneered by Tracy Sonneborn with classic experiments on Paramecium performed more than 40 years ago, and extended more recently to Tetrahymena by Frankel and his colleagues (42, 44). A familiar example is the persistence of ciliary units whose orientation has become reversed during division or conjugation. The explanation follows directly from the manner in which the cells propagate: they elongate, and new ciliary units arise in a definite spatial relationship to an existing unit. Just how ciliary units reproduce is still not well understood, but continuity of the cortex is clearly responsible for their orientation (72).
Even more spectacular, and much harder to explain, is the persistence of a global pattern of cellular organization known as “doublets.” Occasionally the daughters of a dividing cell fail to separate and fuse back to back, producing a doublet cell. The astonishing finding is that doublets propagate indefinitely as doublets; they can even pass through a cyst stage and reemerge as doublets. There is no reason to believe that any alteration of DNA is involved, genetic or epigenetic; the explanation must be that the cortex of the mother cell is continuous with that of its daughters, and some aspects of its structural organization persist across cell division (42). Just what this enduring entity may be remains unknown, one of many unsolved mysteries of cortical continuity.
Ciliates are special in many ways, but the phenomenon of structural inheritance is not confined to them. To mention just one instance, take the selection of the bud site in Saccharomyces cerevisiae (25, 26), to which we shall return below. Localization of the bud depends on cortical landmarks that are laid down during budding, are structurally inherited, and persist at the poles through many budding cycles. The proteins that make up the landmark are, of course, genetically specified; but inheritance of their location is due to the continuity of cell structure.
Things copied.
Genetic information is famously copiable; no other copying process matches the replication of nucleic acids for precision, the capacity for expansion, and universal significance. But several other cell structures undergo some sort of replication, which is part of the way one cell makes another.
Centrosomes are a clear case in point. In the yeast Saccharomyces cerevisiae, the spindle pole body, located on the nuclear membrane, serves as the cell's microtubule-organizing center; it nucleates both the microtubules of the aster and those that make tracks for the transport of the nucleus into the bud. Early in the cell cycle, the single spindle pole body of the interphase cell undergoes duplication; the two offspring position themselves on opposite sides of the elongating nucleus, so that one goes to the bud while the other remains with the mother cell. Just how the spindle pole body duplicates still holds many mysteries (1), but it appears to be a copying process in the sense that an existing body is necessary to make a new one.
Yeast cells have no centrioles, but animal cells and many protists do. Just what centrioles are good for remains enigmatic, though their near identity to the basal bodies of undulipodia suggests some possibilities (12). What concerns us here is that a new centriole normally arises in a definite spatial relationship to an existing one, and at right angles to it. Centriole duplication is part of the mechanism by which the cytoskeleton of the daughter cell is patterned upon that of the mother. It is probably not correct to say that one centriole provides a template to make another (and there are instances of centrioles arising de novo), but some kind of copying appears to be involved.
Finally, note the Golgi. The standard way of doubling the Golgi apparatus, as seen in animal cells, relies on fragmentation into vesicles. These are distributed among the daughter cells and reconstitute the organelle by self-organization. But there are other cases, for instance the protozoan Toxoplasma gondii, in which the Golgi splits transversely “like a pile of paving slabs hit by a karate chop” (115; see also reference 77). A copying process? Perhaps, one that depends somehow on a persistent matrix.
The lesson to be drawn from these and many other observations seems plain enough. As a cell grows, divides, or changes form, it models itself upon itself. New gene products are released into a molecular society that already has spatial structure, and this framework ensures that placement of new molecules is congruent with the old order. To borrow a useful term from Katz (82), the mother cell serves as a templet (not template) for the construction of its daughters, a source of configurational information. Cell heredity, the transmission of characters in ways that do not involve genes, is a by-product of the physiological procedures by which cells transmit spatial architecture to their progeny. But there is a larger point to be made. As far as we know, there is no other way to generate order on the cellular scale: it must be built upon existing order. That is why elements of cell structure, particularly the membranes that define the boundary, have to be listed together with the genes among the features that living systems perpetuate by heredity.
Fields and Gradients
Molecules are nanometer-sized objects organized into cells on the scale of micrometers to millimeters. Can cell growth and development be wholly accounted for by molecular events, by local interactions obeying only local rules? Some sort of spatially extended influence seems to be called for, most probably of a kind traditionally associated with the idea of a field. Physics supplies models, such as electric and gravitational fields extending through space. Embryologists of the early 20th century found the concept of a morphogenetic field useful, and this long-marginalized idea is now reasserting itself in modern dress. A morphogenetic field would be a discrete territory within which genetic information is coherently translated into three-dimensional architecture (48). A gradient of one or more chemical substances, or of some physical condition, would make a plausible candidate for the coordinating agent.
Students of animal development agree that the developing embryo is blocked out by gradients of diffusible molecules (“morphogens”) that supply positional information to the individual cells (53, 94). There is reason to believe that auxin gradients play an analogous role in plant development (13). By contrast, there have been few indications that fields and gradients are involved in localizing organelles or activities within cells themselves, with the singular exception of the ciliates. In these uncommonly large and complex cells, the location of cortical organelles appears to be established in reference to a pair of axes, one longitudinal and one circumferential, that map out a field of positional information; the cell assembles such organelles as the oral apparatus or the contractile vacuole at a particular map location. The remarkable experiments that support and extend this conclusion have been reviewed in detail by Frankel (42-44); unfortunately, both the physical basis of the field and how it functions as a localizing agent have remained utterly refractory.
New evidence for the existence of fields that span cellular space, coordinating and localizing events within it, has come very recently from an unexpected quarter: bacterial cell division. Escherichia coli and other rod-shaped bacteria construct a septum at the midpoint, which they locate with the aid of a set of proteins that oscillate between the cell poles. We shall return to this remarkable discovery in the next segment. Here we note only that the locus of septum construction is specified by the distribution of inhibitory proteins, a clear instance of a field that supplies spatial information.
It seems unlikely that bacteria, the smallest of cells, should be unique in making use of spatial gradients to localize physiological events. Evidence from eukaryotic cells is scarce, but apical growth of fungal hyphae appears to be a promising area. A fungal hypha is a highly polarized filament that directs secretory vesicles to its outermost tip. How does a hypha know where its tip is? The answer is far from certain, but all the hypotheses to be discussed below invoke a spatially extended field of one kind or another.
Force and Form
The shape of every object, whether living or not, is the result of physical forces. Inanimate objects are generally shaped by forces extrinsic to themselves; living organisms, by contrast, are molded by forces of their own making (gravity is an exception). In the case of unicellular organisms, just a few forces account for most of morphogenesis. One class is represented by the weak but numerous noncovalent interactions among molecules, including hydrogen bonds and the association of complementary charges, that shape self-assembling complexes such as ribosomes and viruses. These are clearly insufficient to explain how bacterial rods or fungal hyphae shape themselves, which calls for forces that act on larger aggregates and over longer distances. The two major forces that enter into cell morphogenesis are hydrostatic pressure generated osmotically and mechanical forces exerted by and upon the cytoskeleton.
Hydrostatic pressure, or turgor pressure in walled cells, results from the tendency of water to flow from the dilute medium into the more concentrated cytoplasm. Enlargement of the volume is resisted by the rigid cell wall, and steady state is reached when the internal hydrostatic pressure balances water influx. Turgor pressures of 4 to 5 atmospheres (4 to 5 bars), comparable to that in a bicycle tire, are common in eukaryotic cells and gram-negative bacteria; gram-positive bacteria are more highly pressurized. Turgor presents a problem to growing cells, which must expand their surface area without ever weakening the wall to the point where it may fail. But turgor is also generally believed to be part of the solution: it supplies a force that overcomes both molecular cohesion within the wall itself and external resistance, and thus drives surface expansion. Now, if a cell is to generate any shape other than spherical, surface expansion must be localized. Hydrostatic pressure is a scalar quantity whose magnitude is everywhere the same, and there is no obvious way to localize its application. However, cells can comply with that force in a localized manner by local stretching of the wall and/or by the localized insertion of new wall units. Fifteen years ago, I encapsulated the principle in the mantra that morphogenesis in walled cells results from localized compliance with the global force of hydrostatic pressure (57). The precept is supported by a large body of work from bacteria to higher plants, but it does require restriction and qualification (62).
Localized compliance takes a variety of forms. The thesis that turgor pressure produces the work of wall expansion is central to Arthur Koch's surface stress theory of bacterial morphogenesis (87, 89, 90). The theory accounts for the emergence of cocci, rods, tip-growing filaments, and other shapes by invoking diverse patterns of wall growth. In a few favorable instances, it has even been possible to compute the form a cell should generate. I have been much impressed by the surface stress theory and tried to promote it in my writings. Other reviewers have not shared my enthusiasm (116, 167), and the theory is now under serious challenge from the discovery that bacteria do, after all, possess a cytoskeleton that participates in shaping the cell. Nevertheless, turgor and localized wall expansion remain the most plausible principles to account for bacterial shapes.
Turgor as the driving force for growth remains the conventional wisdom among plant physiologists (27, 134) and perhaps among mycologists too. But the confortable consensus has been challenged by the discovery that certain oömycetes can extend and produce true hyphae even under conditions that reduce turgor to the point where it is no longer measurable (63, 96, 114). Evidently, turgor pressure is not universally required for tip growth. I am inclined to consider this an exceptional situation, possibly an adaptation peculiar to oömycetes, which respond to diminuation of turgor by producing an abnormally plastic wall. Under these conditions, mechanical forces generated as part of the molecular mechanisms of tip growth suffice to drive it forward. If this is correct, the extension of “turgorless” hyphae is similar to cell crawling and draws attention to the second class of morphogenetic forces.
The complement to hydrostatic pressure would be mechanical forces generated at the level of the cytoskeleton; and unlike turgor, mechanical forces can be applied in a localized manner. The most familiar examples are the contractile forces generated when actin filaments slide past one another or past myosin filaments, a process powered by ATP hydrolysis. We are apt to put these under the heading of motility rather than morphogenesis, but every amoeba illustrates that form can be quite a direct reflection of cell movement.
Mechanical forces need not always be generated with the aid of motor proteins. A striking illustration comes from the recent solution to the long-standing problem of how cells crawl. Fibroblasts, neutrophils, and other crawling cells advance by protruding a flat, seemingly structureless protrusion called a lamellipodium. What pushes the lamellipodium forward? Not, it now appears, motor proteins such as myosin; the key is the assembly and polymerization of actin. Current opinion holds (124, 133) that the cells assemble a three-dimensional meshwork of actin filaments (an ATP-driven process), which push the plasma membrane outward and thus make the lamellipodium protrude. This, I suspect, is also a feature of apical growth in fungal hyphae, as displayed by the extension of “turgorless” oömycete hyphae. In both cases, we see morphogenesis that results from the localized application of mechanical force.
The hierarchy of order constitutes, I believe, a general and comprehensible answer to the question of how molecules come together to make cells. The essential point is that this cannot be accomplished by a unitary, universal mechanism, analogous to the way amino acids join to make proteins. Making a cell requires a succession of stepping stones which collectively and progressively bridge the gap between nanometer-sized molecules and cells three to six orders of magnitude larger. There is nothing fundamental about the particular stepping stones set out above. Each one is, of course, drawn from the facts of nature, but their arrangement is intended only to assist the imagination, and the path must be judged by its utility in helping the mind to cross. Walk with care and mind the gap.
GROWING JUST SO
All levels of the hierarchy of order come together in the study of cell morphogenesis, which seeks to render a causal and dynamic account of both form and organization. In no instance has that objective been reached, but enough is known to illustrate how the stepping stones laid in the preceding segment apply in practice. A conspicuous cleavage runs between eubacteria and eukaryotes, each representing a distinct pattern of cellular order. The essential difference may be simply one of size: the eukaryotic way allows cells to grow much larger than the bacterial one, although not all eukaryotic cells avail themselves of that option. Archaea represent a third pattern of order, but this is more noticeable at the molecular level than at the cellular one; archaea will therefore not be considered here. I have selected examples from the eubacterial and eukaryotic domains to illustrate some of the ways in which cells establish location, direction, and overall form. The treatment purposely omits almost all detail so as to highlight those aspects that pertain to the organization of events in cell space.
Escherichia coli: Bisecting the Cylinder
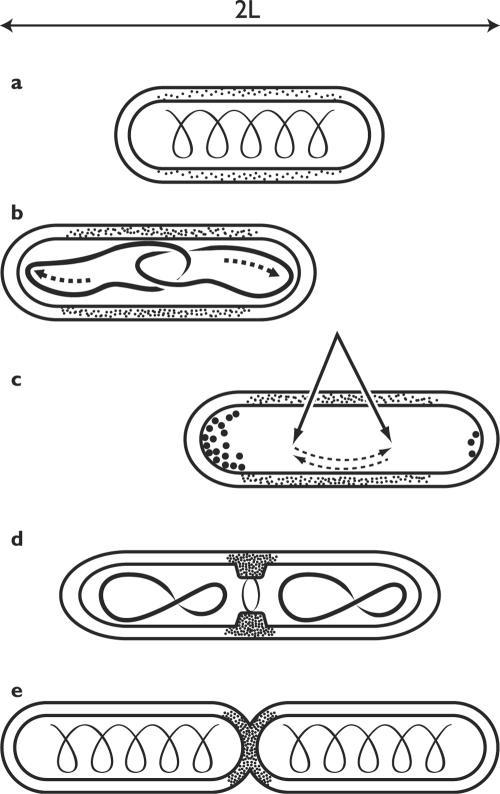
A bacterial rod, such as E. coli or Bacillus subtilis, can be abstractly represented as a cylinder with rounded caps, 2 to 5 μm in length. Tiny as it is, each cell still contains some 50 million molecules large and small, not counting ions and water (119). The majority are probably free to diffuse, but some occupy a definite address in cell space, at least transiently, and these play the key roles in the reproduction of the system. Growth and division proceed in a carefully choreographed sequence of steps that overlap in time and space (Fig. 4): biosynthesis of metabolites and macromolecules; duplication of the genome and segregation of the products; elongation of the cell at constant diameter to the proper length; finding the cell's midpoint; construction of a septum; and finally, cleavage of the septum and separation of the daughters. The ballet generates two cells, each a cylinder with rounded caps exactly like the original one. Bacterial division is presently an intensely active field and the subject of numerous excellent reviews (40, 46, 105, 117, 139, 140).
FIG. 4.
Duplicating the rod: some localized and oriented processes. (a) Actin-like cytoskeleton and dispersed synthesis of sidewall. Stippling indicates intensity of wall synthesis. (b) Segregation of nucleoids to the poles by an active mechanism; sidewalls elongate. (c) Cell finds its midpoint by the oscillation of Min proteins (nucleoid occlusion not shown). (d) Construction of divisome at the midpoint, wall synthesis focused there. (e) Construction of the septum, sidewall elongation ceases. Note that the cytoskeleton presumably undergoes rearrangements during the cell cycle which remain to be described.
Not so long ago, bacterial physiologists assumed that DNA polymerase is free in the cytoplasm and travels along the chromosome as it replicates the genome. That was never very plausible and now appears to be untrue. In Bacillus subtilis and E. coli, at least, DNA polymerase and accessory proteins are fixed to the cell envelope near its middle, and DNA is replicated by threading it through the “replication factory” (46). In both organisms DNA replication must take place in an oriented manner, for the newly duplicated origins become attached to the cell poles; a specific marker or tethering protein must be located there. How are the duplicated DNA threads, whose unraveled length would be a thousand times greater than that of the entire cell, pulled apart and segregated? There is reason to believe that drastic condensation of DNA supplies part of the answer. But there is also evidence for the participation of some kind of mitotic machinery that exerts mechanical force and pulls the nascent chromosomes apart (46); the nature of this machinery is still quite uncertain. In the end, the chromosome comes to be organized into loops perpendicular to the cell axis, with individual genes occupying fixed positions in linear order between origin and terminus (14, 156). The compulsive neatness of the arrangements is a far cry from the historical image of DNA tangled up in midcell like spaghetti in a bowl! One cannot help wondering whether there is, after all, a spatial relationship between the position of genes and the location of their products in cell space (28, 116).
Division of rod-shaped cells turns on the construction of a septum that bisects the cylinder precisely in the middle. How cells find their midpoint and direct the synthesis of new cell wall to that locus is currently being clarified and appears to involve unexpected and novel mechanisms of dynamic self-assembly and cellular self-organization. A major advance stemmed from the discovery that the protein FtsZ, known from mutants to play an essential role early in septum construction, is localized to a ring at the midpoint of dividing E. coli cells (100). FtsZ proved to be a homolog of eukaryotic β-tubulin and, like tubulin, a GTP-hydrolyzing enzyme. Several other proteins, likewise first identified in screens for mutants defective in division, contribute to the assembly of the Z-ring; one of these, ZipA, is thought to anchor the ring to the cell envelope. It should be noted that the Z-ring is a dynamic structure; only about a third of the cell's complement of FtsZ is found in the ring, and the bound protein exchanges continuously with that free in the cytoplasm. Once assembly is complete, the ring constricts; as it does so, it pulls inward on the plasma membrane and cell envelope and is thought to reposition the particular peptidoglycan synthase required to produce the septum. Just how the Z-ring assembles and precisely how it constricts and how peptidoglycan synthesis is redirected from the sidewalls to the septum all remain matters for speculation and debate (reviewed in references 40 and 117). Nanninga (117) has proposed the useful term divisome to designate this dynamic machine, probably self-constructing, that appears to consume metabolic energy (GTP) to support the work of cleaving the cytoplasm (Fig. 4). In doing so, the divisome will be working counter to the force of turgor pressure that tends to expand the cell surface.
FtsZ and closely related homologs represent the prokaryotic way to divide; they are nearly ubiquitous among eubacteria and archaea. But there are exceptions: FtsZ is absent from chlamydia and from the crenarchaea. It is found in chloroplasts, but absent from most mitochondria (40, 105). The other divisome proteins are quite irregular in their distribution: FtsA, for example, a prominent component in both E. coli and Bacillus subtilis, is absent from mycobacteria, cyanobacteria, and archaea (105). Evidently, it is possible to construct what appears to be, physiologically speaking, the same machinery from different components.
So far so good. But how does the cell identify its midpoint so as to localize the septum correctly? That has been one of the perennial mysteries of bacterial physiology, and to see it being solved at last is immensely satisfying. Nearly 40 years ago, Adler and his associates isolated a class of mutants that divide aberrantly: instead of locating the septum at the cell center, they tend to place it at one end, producing “minicells” devoid of DNA. The min locus contains several genes whose products map cell space; their task is not so much to promote septum construction at the right place as to block construction at the silent sites adjacent to the poles.
It is truly a remarkable story (reviewed in references 40, 49, 139, 140, and 147). The heart of the matter is that a complex of the three Min proteins (C, D, and E) assembles in one half of the cell, blocking assembly of the divisome there. The complex then disassembles and reforms in the other half of the cell. The oscillations repeat with a period of about 20 seconds. The crucial element is probably MinE, which serves as a mobile cap that sweeps MinC and MinD first to one pole and then to the other. The molecular details are beyond my scope here, except to take note of the recent discovery (148) that the Min proteins are not free to wander but are organized into extended coiled structures that wind around the cell from pole to pole beneath the plasma membrane. And how would oscillations in the localization of the Min proteins identify the cell's midpoint? The operation of a windshield wiper offers a clue: the midpoint is where the time-averaged concentration of the inhibitory protein MinCD is lowest, allowing assembly of the divisome to begin there (56).
Powerful support for the conception as a whole comes from computer simulation. Meinhardt and de Boer (112) have modeled pole-to-pole oscillations of the Min proteins and found that they can indeed localize the cell's midpoint. The process is entirely self-organizing and requires no existing topological markers; it generates a field over which MinC, MinD, and MinE oscillate spontaneously, with concentrations lowest in the center of the field. The model will now have to be revised to accommodate the finding that Min proteins are moving within some sort of framework (148). Besides, when the whole story is told at last, it will have at least two additional episodes. One will recount the role of the nucleoids, which somehow contribute to blocking septum assembly in adjacent regions of the membrane (117). The other will deal with variations on the theme of finding the middle: both Bacillus subtilis and Caulobacter crescentus do it without traveling waves of MinE (cited in references 49 and 112).
All the while that the cell has been replicating its genome and preparing to divide, it has also been doubling in length. That entails the deposition of new envelope material along the cell's sidewalls, particularly of peptidoglycan, which confers upon the wall mechanical strength and overall shape. Sidewalls and poles are chemically indistinguishable but metabolically dissimilar. Poles, once deposition of the septum has been completed, are nearly inert; sidewalls, by contrast, undergo turnover even while incorporating fresh peptidoglycan, Since the peptidoglycan layer is extensively cross-linked, extension requires cutting the fabric and splicing new wall units into place. The mechanism has recently been explained by Höltje (69), who postulates a smart synthase that splits out one old strand while inserting three new ones in its place. It is important that wall growth in rod-shaped bacteria be dispersed: new units are inserted all along the length of the cell. How that is arranged is not yet certain, but there is strong evidence that those recently discovered actinlike cytoskeleton filaments are required to ensure this pattern of wall syntheses (29, 40). One can imagine a helical scaffold, studded with peptidoglycan synthase, sweeping over the wall's inner surface as it rotates upon the cell's axis.
Now, how would all this localized and directional biochemistry produce a cylinder with rounded caps? The poles are not hard to understand, at least in principle: poles form by lengthwise cleavage of the septum and are then bowed out by the force of turgor pressure. The degree of stretching is a function of the chemical structure of the cell's particular peptidoglycan. But what shapes the cylinder, with its smooth sidewalls and constant diameter? For the past two decades, the only general answer to this question has come from Arthur Koch's surface stress theory (87, 89, 90). In his view, the force that drives expansion of the surface is hydrostatic pressure, and the cell complies with that force by the insertion of new units into the wall. The shape of the resulting structure is wholly determined by physics, once biology has supplied parameters such as fixed poles, the pressure, a term analogous to surface tension, and the dispersed pattern of wall enlargement. Remarkably, if the critical parameters fall into the correct range, physical theory predicts that a cylindrical form can be produced. E. coli, with its very thin peptidoglycan layer, presents some special issues, but in more tractable cases it is possible to calculate the shape that the cell will assume.
So is it the case that the cylindrical shape of E. coli, like that of a soap bubble carefully inflated between two fixed supports, is generated by physical forces alone? I think the answer emerging from the laboratories is no, and the surface stress theory will have to be very significantly amended to bring it into concord with the data. It remains true that turgor pressure is the dominant (or even the sole) force driving surface expansion and that the shapes of bacteria result from the manner in which the cells comply with that global force by the insertion of new units into the wall. In some cases, notably the streptococci, surface stress seems sufficient to account for the cell's form (91). But in E. coli and other rod-shaped bacteria, the large number of mutants that display aberrant forms clearly indicates that specific proteins are required to ensure the constancy of cell diameter and length, not to mention cytokinesis.
The cytoskeleton is deeply engaged in morphogenesis: it localizes wall synthesis, may supply mechanical support, and also helps do the work (4, 5, 29, 49, 78, 167). Bacterial forms are produced by the interplay of at least two kinds of force: turgor pressure, the global force pressing to enlarge the cell, and local forces generated by localized molecular machines such as the divisome. We have some way to go, but when the dust settles we should be significantly closer to a satisfying answer to that innocent question: Daddy, how does E. coli grow a cylinder?
Caulobacter crescentus: Organizing the Poles
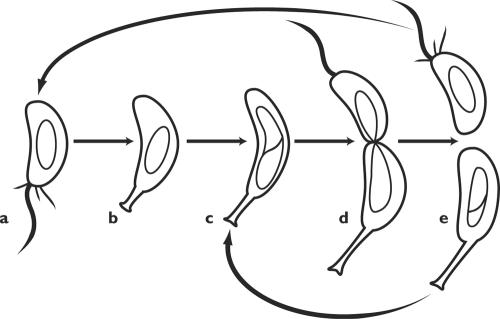
The life cycle of caulobacteria, unlike that of E. coli, is visibly polarized. The crescent-shaped cells grow attached to a surface by a stalk, which may be as much as 20 times the length of the cell body. The stalk is continuous with the cytoplasm but devoid of ribosomes and DNA; it probably assists in solute uptake by greatly increasing the cell's surface area, which allows the organisms to flourish in sparse medium. The growing cell elongates and divides into two dissimilar daughters. The proximal one is sessile; it remains attached to the stalk and continues to grow and divide. The distal “swarmer” cell is mobile and swims off in search of a livable home; when it settles down, it generates a new stalk and begins the cycle anew (Fig. 5).
FIG. 5.
The life cycle of Caulobacter crescentus. (a) Motile swarmer cell. (b) The swarmer cell settles down, loses its flagellum and pili and forms a stalk in their place. (c) As the cell grows, it begins to replicate DNA and assembles flagellar precursors at the distal pole. (d) Prior to division, the distal pole sprouts a flagellum and motility is activated. (e) The progeny stalked cell initiates a new round of replication, while the progeny swarmer cell swims off. (After Ausmees and Jacobs-Wagner [4], with permission of the publisher.)
The polarity of the life cycle is made manifest by the cell's appendages. The new poles generated by cell division are bald. As the stalked daughter cell elongates, and well before the septum divides it in two, the distal pole sprouts a single flagellum, a cluster of pili, and an aggregate of chemotaxis receptor proteins. These pass to the swarmer daughter; when that, in turn, settles down, all those appendages are dismantled, the flagellum is ejected into the medium, and a new stalk forms in its place. The Caulobacter cell cycle thus displays features more commonly seen in eukaryotes: differentiation within a polarized cell, followed by asymmetric division producing progeny that differ in form and behavior. The developmental biology of C. crescentus is being intensely studied in several laboratories and is the subject of frequent reviews (4, 19, 38, 110, 146, 147). Among the themes that emerge from the welter of data are the importance of localizing signaling proteins and the pivotal role of the cell poles in generating spatial order.
The two daughter cells produced upon division contain the same genome but express different genes and exhibit different physiologies. One glaring example is that the stalked daughter immediately initiates a new round of DNA replication and begins a fresh cycle; by contrast, in the swarmer cell, DNA replication is blocked and remains so until that cell has settled and put forth a stalk. Can one account for polarized development as a consequence of differential gene expression? No, but spatial and temporal regulation of gene expression is part of the story. There is an overall correlation between the time of transcription of a particular gene and the time its product is needed (“just-in-time delivery”). There is also good evidence that, following chromosome duplication but well before septum closure, a barrier comes to divide the elongating cell into distinct compartments (80); the nature of the barrier is still uncertain, but it is presumed to be an outgrowth of the plasma membrane. Some genes are expressed solely in one compartment or the other. Among these are genes required in the final stage of flagellar assembly, building the filament, which only become active in the incipient swarmer compartment.
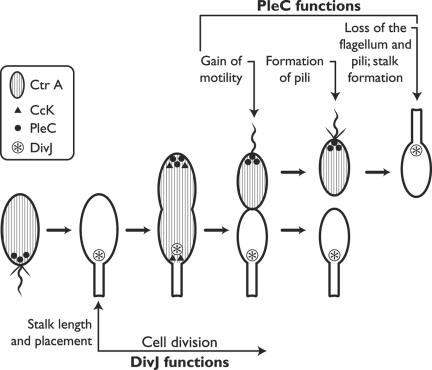
Differential gene expression is itself a consequence of the differential distribution of regulatory molecules (Fig. 6). A particularly important one is the master regulator CtrA, which controls the expression of nearly 100 genes linked to the cell cycle. The phosphorylated form of CtrA, CtrA-P, which blocks the initiation of DNA replication and also promotes the expression of flagellar genes, is dispersed throughout the early predivisional cell. In due course it comes to be restricted to the incipient swarmer compartment; its absence from the incipient stalked compartment is due to several localized degradative reactions, including both dephosphorylation and proteolysis (142). So the spatial regulation of gene expression is one aspect of polarized development, but a relatively late one. Other important stages are represented by the targeting of specific proteins to critical locations in cell space.
FIG. 6.
Localization of some signal transduction proteins during the cell cycle of Caulobacter crescentus. See text for the functions of the kinases. (Adapted from Ausmees and Jacobs-Wagner [4] with permission of the publisher.)
The single flagellum that will confer motility upon the newborn swarmer cell is a conspicuous feature of the distal pole, and its assembly starts early. Some 50 genes are required to specify the parts; they are transcribed sequentially, and their protein products are assembled in approximately the same order (19, 38). The MS ring, embedded in the plasma membrane, comes first and provides a platform for the subsequent assembly of the motor, the hook, and eventually the filament. How does the elongating cell “know” where its pole is? The actinlike cytoskeletal protein MreB evidently plays a major role in establishing cell polarity (49, 50), probably assisted by positional markers left at the pole when it formed during the preceding cell cycle (Fig. 5). Cell wall carbohydrates, lipid domains, and specific proteins could all serve as positional markers and provide anchorage to the protein(s) that initiates assembly of the flagellum (4, 101). The chemical nature of these markers, and just how they and the flagellar proteins travel to the construction site, are key questions for the future.
Preparations at the pole include localizing there a set of regulatory proteins of the kind known as two-component systems: a sensory element which, in response to some input, phosphorylates a response regulator that effects the response (transcription of a particular set of genes, perhaps). The master regulator CtrA-P, mentioned above, is a case in point. That protein is dispersed in the cytoplasm, but the sensory kinase that phosphorylates CtrA, designated CckA, is localized to the cell poles early in the cell cycle. A plausible interpretation holds that CckA monitors some aspect of the cycle and that it is most active when aggregated at the cell poles (146, 147). Later on, as the swarmer cell settles and prepares to synthesize a stalk, CtrA itself becomes bound to the cell pole just prior to its degradation (142).
Pili form at the same distal pole but are regulated independently. Here again a protein kinase is involved, called PleC, which controls localization of a special secretory apparatus but not the assembly of flagella. This kinase is localized to the flagellar pole in both the predivisional and the swarmer cell but is cleared out before stalk emergence. In this case, an anchor protein has recently been found: PodJ supplies positional information that localizes PleC to the flagellar pole (155). Just how PodJ does this and how it comes to be positioned at the pole remain to be discovered. A fourth protein kinase, DivJ, localizes to the stalked pole and plays an essential role in stalk placement and in cell cycle progression.
Chemotaxis receptors are yet another polar feature, with their own mode of localization. The proteins are synthesized in the predivisional cell and then segregated into the swarmer compartment. This requires a particular motif of 14 amino acids, whose function may be to recognize a positional marker or anchor. Several other members of the chemotaxis cascade must also be present. How the receptors travel to the pole and what keeps them there are not yet clear.
Step by step, advancing systematically from the bottom up, the molecular mechanics of polarized development in C. crescentus are being worked out. Though still fragmentary, the story is already exceedingly complicated, not easily grasped as a causal, coherent whole even with the aid of a wiring diagram (110). As more information accumulates, the diagram may have to be expanded into three or even four dimensions. Keep in mind also that this particular web is more or less unique; E. coli is the product of another, rather different web. All the same, I believe that one can extract some general features that may apply to prokaryotic cells generally.
i. Progression through the cell cycle depends on the controlled expression of genes, ordered both temporally and spatially. Gene expression, in turn, is monitored by checkpoints which ensure that morphogenesis and transcription stay in register.
ii. Production of functional proteins is tightly controlled, so that they are made when required and degraded once their job is done.
iii. Location of a protein may control its activity: not only must catalysts be present, they must be present at the right place.
iv. Proteins become localized by binding to a positional marker and perhaps by other mechanisms yet to be discovered. They probably move around the cell by diffusion, either in the cytoplasm or in the membrane, followed by capture and retention. Thus far, there is no unambiguous evidence for motor proteins or for targeting.
v. Localized proteins display, execute, and help to maintain polarity, but no single one is its cause. Asymmetry is present continually, a feature of the cellular system as a whole. The crucial step that regenerates asymmetry at each division is the formation of two fresh cell poles.
Apical Growth: a Focus for Secretion
What do fungal hyphae, budding yeast, germinating spores, and pollen tubes have in common? They are all walled, eukaryotic cells that grow at a tip or apex. In walled cells, morphogenesis turns on the pattern of wall synthesis; apical growth ensues when the deposition of new wall is confined to a small region, which advances and becomes the tip. A century of research on apical growth (reviewed in reference 66) has given rise to a number of general insights. First, apical growth is not a matter of wall chemistry; the forms of chitinous hyphae and cellulosic ones differ only in detail. Second, apical growth results from the polarized and localized secretion of vesicles carrying wall precursors; exocytosis is confined to the tip of the tip. Third, the cytoskeleton is key to making the tip. Fourth, morphogenesis can be envisaged as an instance of local compliance with global force: turgor pressure drives expansion, localized secretion lets the wall yield locally, and wall properties shape the tip. Last but not least, there are several ways to focus secretion so as to grow a tip (Fig. 7).
FIG. 7.
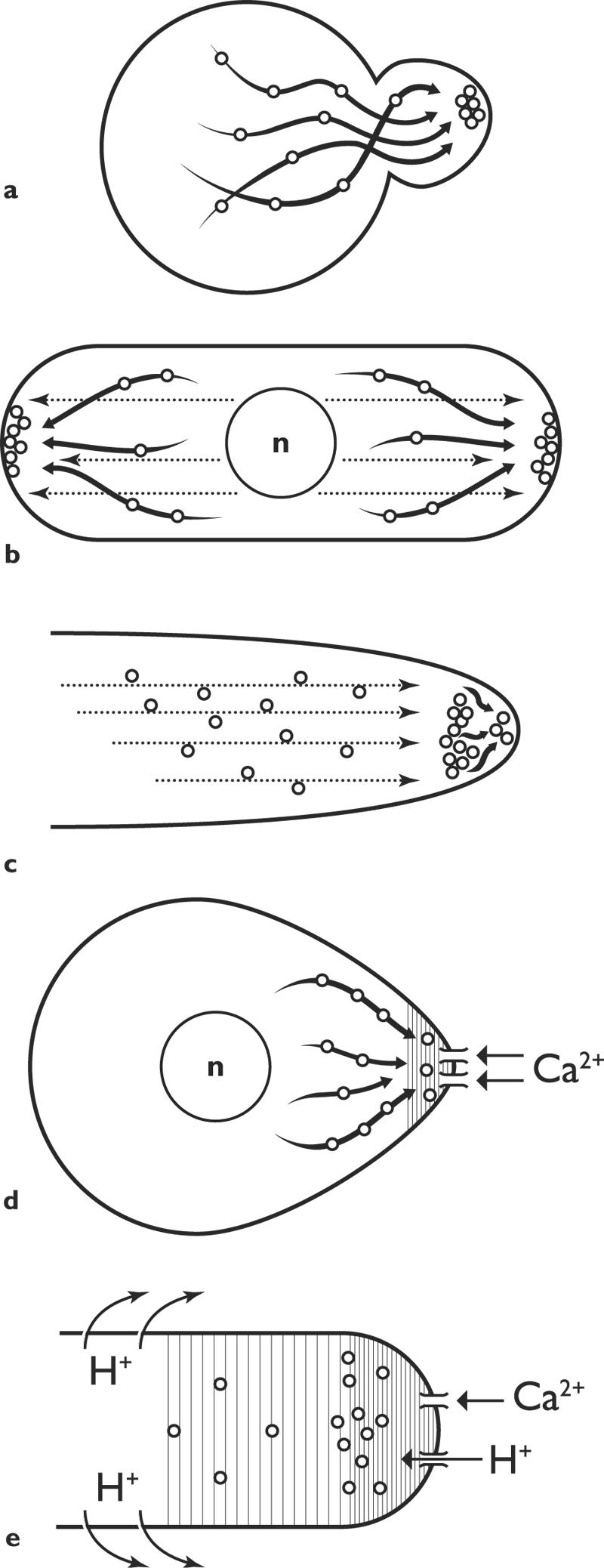
Targeting secretory vesicles: some current ideas. a. Saccharomyces cerevisiae: vesicles delivered to marked site on actin cables. (After Pruyne and Bretsher [131], with permission of the publisher.) b. Schizosaccharomyces pombe: vesicles delivered on actin cables to apical sites, marked by proteins carried on microtubules. c. Fungal hyphae: vesicles delivered on microtubules to a Spitzenkörper, from which they proceed to the apex, perhaps via actin filaments. d. Silvetia compressa (previously known as Pelvetia compressa): a current of calcium ions localizes the site of outgrowth, to which secretory vesicles travel on actin cables. e. Lily pollen tube: currents of calcium ions and protons into the tip localize the site of exocytosis; actin involved in this and in vesicle transport.
Saccharomyces cerevisiae.
We begin with baker's yeast, not because budding is such a notable instance of apical growth but because yeast supplies the paradigm to which all other organisms must be compared. The heart of the matter is the establishment of a vectorial secretory pathway that delivers Golgi vesicles, laden with enzymes and wall precursors, to the site of construction and also confines their exocytosis to a chosen location (Fig. 7a). The form of the bud emerges, presumably in compliance with turgor pressure, as the target locus shifts over time in an orderly sequence. Many open questions remain concerning the nature of those vesicles, just what they contain, and just how new wall units (glucan, in this instance) are incorporated into the existing wall. What has now been established is that the vesicles travel along cytoplasmic cables composed of actin, tropomyosin, and auxiliary proteins, powered by a particular myosin (Myo2p) and ATP. The cables deliver the vesicles to the site of exocytosis, or at least very close to it (15, 131, 132). Actin cables and Myo2p also participate in the segregation of the vacuole and the positioning of nucleus; they, rather than microtubules, organize the cytoplasm of yeast cells.
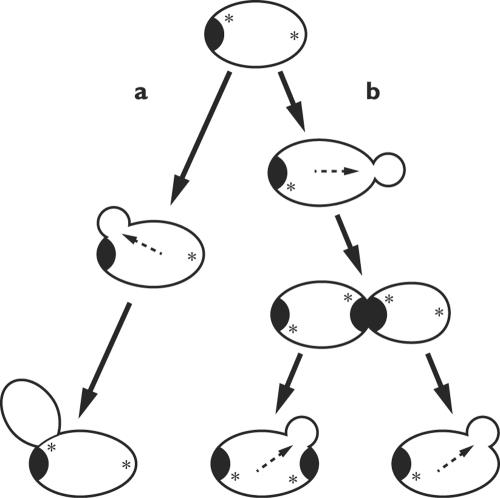
The vectorial secretory apparatus defines the axis along which cell division becomes organized; how is it put in place? Polarized morphogenesis in yeast has been neatly construed as a hierarchy of four consecutive steps, beginning locally but turning progressively global (25, 128). First, the cell determines a bud site on its surface, reading either endogenous cues generated by the cell itself or exogenous ones from the environment. This site is then marked by the deposition of a landmark, or positional marker, in the cell cortex (Fig. 8). In the third step, activation of a signal cascade initiates polarization of the actin cytoskeleton upon the landmark. Finally, the secretory apparatus and other functions are organized around the cascade, and polarized growth begins. Note that the secretory apparatus is constructed from the periphery inward, not from the nucleus outward.
FIG. 8.
Cortical landmark localizes the site of the next bud in diploid yeast cells. Diploid cells bud from the poles, either the pole that bears the birth scar (a) or its opposite (b). Localization depends on cortical landmarks (*) laid down during budding and transmitted structurally. Internal arrows indicate the axis of polarization. (Based on studies by Chant [26] and others.)
Polarized growth requires a large number of proteins, a hundred or more, whose functions and interactions are receiving much attention (25, 26, 33, 102, 131). Among their tasks are to specify bud location and also the signaling cascade centering on that key player, the GTPase Cdc42; to organize the polar cap and the actin cables that reach deep into the mother cell; and to generate the septin rings that define membrane compartments for the bud and the mother and subsequently provide a platform for the construction of the septum. Some are part of the complex known as the exocyst, which governs the docking and fusion of secretory vesicles (153). These proteins make up the obligatory nuts and bolts, but definition of an axis of polarity takes place not on a biochemical level but on the cellular one.
In S. cerevisiae, the budding pattern is normally quite stereotyped. Haploid cells bud adjacent to the previous bud, and diploid ones bud from either end. These sites are marked by the deposition of marker proteins, specified by various BUD genes, at previous sites of division or growth (25, 26). These landmarks pass from one generation to the next by structural inheritance, a consequence of cell continuity (Fig. 8). But these inherited cell vectors represent a bias, not a command; cells can dispense with them or override them. For example, mutants deficient in the BUD genes can still construct buds, only they do so at random locations. The explanation may reside in the spontaneous amplification of a local fluctuation in the activity of Cdc42, the GTPase that unleashes all subsequent events (158). In other cases the promise of sex overcomes normal inhibitions: a gradient of mating pheromone overrules the signals of the bud proteins, reorienting the secretory machinery to produce a “shmoo” pointed towards the prospective partner. Causality is circular: if proteins by their interactions organize the cell, it is no less true that the cell organizes its proteins.
Fission yeast.
Schizosaccharomyces pombe resembles its distant cousin S. cerevisiae in growing at an apex, but it does so in its own fashion. Newborn cells are cylindrical with rounded ends, 3 to 4 μm in diameter and about 8 μm in length, with the nucleus in the middle. They grow by elongation, first at one end only and then at both, keeping a straight axis. When the length has doubled elongation ceases, mitosis and cytokinesis ensue, and a septum bisects the cylinder (Fig. 7b). Note how different this pattern is from that of E. coli, which generates much the same form: the bacterium elongates the side walls while the poles are inert; Schizosaccharomyces pombe grows apically, at the poles. Schizosaccharomyces pombe is becoming a favorite model organism for research on cell morphogenesis, frequently reviewed (20, 24, 25, 65).
We don't actually know how S. pombe constructs the apical wall, but from the distribution of actin cables and actin patches during the cell cycle (106, 126), one can infer a pattern very similar to that described above for budding yeast: secretory vesicles transported along actin cables, to be exocytosed at sites of wall construction such as the growing tip and septum. The exocyst is required for some secretory processes, but apparently not for all (157). Clear differences appear at the level of the microtubule cytoskeleton (55): unlike Saccharomyces cerevisiae, which makes almost no use of microtubules for purposes of cell polarity, Schizosaccharomyces pombe relies on them to keep its axis straight.
Briefly, mutants deficient in organizing the microtubule cytoskeleton tend to produce cells that are T-shaped (branched), reflecting the emergence of a new growth axis. These and other defects are especially pronounced in mutants deficient in a protein designated Tea (for tip elongation aberrant). Tea1p is carried upon microtubules to the cell apices, where it is deposited. Neither microtubules nor Tea1p is absolutely required for polarized extension, which can proceed even in their absence; and just what Tea1p does is still rather uncertain. A recent model (143) suggests that Tea1p and microtubules are required to establish polarity after division or to reestablish it after disruption. Tea1p, targeted to the tip by longitudinal microtubules, helps to recruit the actin-based secretory machinery to the poles and may itself be an integral component of the landmark that identifies the pole. It seems, at least for the present, to be a unique mechanism of specifying positional information.
Fungal hyphae.
Growing straight and narrow for millimeters and more, hyphae are the epitome of apical growth. Each hypha is an intensely polarized secretory system. Vesicles, laden with precursors and enzymes for the construction of wall and plasma membrane, are manufactured in Golgi bodies along the trunk and brought forward; they fill the hyphal apex and undergo exocytosis at its tip. Deposition of new wall is confined to that apex and is so organized as to generate a tube of constant diameter capped by a tapered tip that is continuously made afresh as it advances (Fig. 7c). In the canonical view (57, 62, 67, 88, 159, 160) the nascent wall deposited at the tip is plastic but hardens within seconds to minutes as it falls behind the advancing tip and joins the trunk. Plastic wall yields locally to forces exerted upon it, the chief of which will be turgor pressure; this is what shapes the tip and makes it advance.
The question for us is how the apical region is organized so as to focus exocytosis on the extreme tip. Despite much recent discovery and reflection, many of the most basic questions have only partial answers or none at all. What we need most is a set of well-articulated hypotheses to serve as touchstones by which to assess the meaning of data old and new. Plausible ideas will connect with the yeast paradigm but must go farther: hyphae grow continually and faster, in a less stereotyped manner, and with the production of branches (64). Two general ideas are currently under discussion, one centering on the Spitzenkörper and the other on calcium gradients.
Hyphal tips typically display an ephemeral organelle, the Spitzenkörper or apical body, which is present so long as the hypha extends and vanishes when it stops. Ultrastructural studies have revealed a dense aggregate of vesicles surrounding a core containing actin filaments. According to the hypothesis developed by S. Bartnicki-Garcia and G. Gierz, this body is the key to how hyphae grow: it serves as the immediate donor of vesicles for the construction of apical wall and membrane, and its movements determine the direction of extension and the shape of the tip (8, 9, 11, 59). The cardinal virtue of this proposal is that an abstract version can be modeled mathematically, generating a predicted shape that is uncannily like a fungal hypha in cross-section. Extension to three dimensions makes the mathematics more demanding and requires one to specify parameters beyond those needed for two dimensions but yields satisfying forms (10, 47). The hypothesis has been extensively tested by experiments to document that manipulation of the Spitzenkörper alters tip shape and growth direction in the manner predicted by the model (135, 136). Many aspects need to be specified: just how do secretory vesicles travel to the Spitzenkörper and then onward? What happens to them there? How are the movements of that transient organelle coupled to extension of the cytoskeleton? All the same, in my opinion Bartnicki-Garcia and his colleagues are looking in the right direction; brutally abstract as it is, their model has the ring of truth.
The only substantial alternative credits the influx of calcium ions into the tip with setting up a localized ion gradient that regulates exocytosis and maps out the tip. In effect, calcium ions serve as a morphogen. Historically, this idea goes back 30 years to the pioneering studies of L. F. Jaffe, R. Nuccitelli, and K. R. Robinson on the germination of fertilized eggs of the brown alga genera Fucus and Silvetia (Fig. 7d), and it continues to resonate in that field (92, 137). In recent years, calcium gradients have become prominent in attempts to make sense of apical growth in organisms that lack a Spitzenkörper, such as oömycetes and pollen tubes, but they are also turning up in the physiology of orthodox fungi.
The proposal states that calcium ions flow into the extending tip through short-lived, stretch-activated channels carried to the tip on secretory vesicles. Calcium ions stimulate local exocytosis in a concentration-dependent manner, either directly or more likely via their influence on actin dynamics. There is compelling evidence from extensive studies on pollen tubes that a calcium gradient exists (Fig. 7e) and plays the kind of role sketched above; but I hasten to add that reality is rather more complicated and poses as many riddles as it solves (41, 68). Calcium gradients, with concentration maximal at the tip, are also present in the hyphae of oömycetes and true fungi; they play a critical role in organizing the tip, but perhaps not in the manner outlined here (149, 150, 154). Even if I were to review the data in depth, I fear that the interpretation would be murky. Nothing would so sharpen our perception as a mathematical model that makes specific and testable predictions.
Molecular optimists may have anticipated that a clearer understanding of hyphal extension would emerge from completion of the genomes of Neurospora crassa and Aspergillus nidulans and from a search for homologs of the pertinent yeast genes. In the event, as Harris and Momany (64) show in a most useful review, matters are very much more complicated. Genes for certain core building blocks are probably universal: actin, tubulins, myosins, and kinesins underpin hyphal growth just as they do yeast morphogenesis. Genes homologous to those of the yeast polarisome are conserved in Aspergillus nidulans, and their products presumably organize the assembly of actin filaments at the tip. But the distribution of so familiar an actor as Cdc42 is spotty; it is needed for the emergence of germ tubes from spores, but is probably not essential for hyphal extension (an unrelated GTPase may take its place). Besides, there is the question of size. Hyphae must specify spatial order on a grander scale than Saccharomyces cerevisiae, and the evidence suggests that they employ gradients and fields that may have no parallel in yeast cells.
PUTTING THE CELL BACK IN THE CENTER
The previous two segments spelled out a rational conception of how molecules come together to make cells and illustrated its experimental foundations with examples drawn from both prokaryotic and eukaryotic microbes. This final segment seeks to extract wider meaning from the science and therefore ventures beyond the laboratory door into a zone of personal judgment and opinion. As a first step, let me briefly summarize what, in my view, we should infer from the experiments and observations surveyed above.
Cells Make Themselves
The spatial organization of cells, including the arrangement of cytoplasmic constituents and the cells' global form, is not explicitly spelled out in the genome. Genes specify only the primary sequences of macromolecules, portions of which are indeed relevant to the localization of those molecules in space. But cell architecture, for the most part, arises epigenetically from the interactions of numerous gene products. Many of these interactions can be well described as instances of molecular self-organization, either self-assembly or dynamic self-construction. However, when self-organizing chemistry takes place in a living cell, it comes under guidance and constraint from the cellular system as a whole. New gene products, and also the output of biosynthesis, are never altogether at liberty; they are released into a cellular milieu that already possesses spatial structure, and they find their places in that pattern under the influence of the existing order. Governing features include cellular compartments and vectors, spatial markers of various sorts, heritable membranes, tracks and channels for intracellular transport, gradients that supply positional information, and an array of physical forces. In eukaryotic cells, and probably in prokaryotic ones as well, the cytoskeleton is the chief instrument for creating cellular architecture and is itself a product of its own operations. Every cell is continuous with its progenitor cell, not only genetically but also structurally; and the new cell will, in turn, serve as a templet (a source of configurational information) for the construction of its own daughters. For that reason, I argue that the smallest entity that can be truly said to organize itself is the whole cell.
That is all well and good, some readers may say, but it sidesteps the question posed at the beginning of this essay. Whence comes the “information” that specifies a cell's spatial organization? Where do we find the “instructions” that make E. coli a short rod or arrange the yeast cytoskeleton and ensure the faithful propagation of those forms? There appears to be a presumption, seldom articulated but taken for granted, that a subset of the genome must specify the higher levels of order and that these genes embody a hard-wired genetic program which actively controls and directs growth and development. The terminology of a program seems quite apt for the cascade of transcription factors that progressively establish cellular identities during the development of an embryo (3). But there is no sign that an analogous network of genes maps out spatial organization at the level of single cells. Note especially that nothing resembling a program for morphogenesis can be identified in the many microbial genomes now on record.
Genes do contribute to cell architecture by way of sequences that govern the association of macromolecules one with another or direct gene products to particular targets. But genes do not encode large-scale order; as Britten (18) put it, “There are no known genes that individually encode large amounts of information specifying the structure or patterns of development.” There is also no reason to believe that, as Penman (125) surmised a decade ago, spatial specifications might be concealed in the vast stretches of noncoding DNA. No, the question was misphrased to begin with. Unlike sequence information, which is encoded in linear texts that are copied, conserved, and translated, architectural order is transitory and emergent; it comes and goes (think of the association and dissociation of subunits). Spatial order is not encoded anywhere at all but emerges from the interactions of the cell's molecular building blocks; it arises by self-organization, like the specifications of a termite mound or the unique jumble of streets in my home town of Seattle. And the propagation of order down the generations depends not on a codebook but on history repeating itself: the same building blocks, released into the same constraining context, will reproduce the same structure time and again.
A Salute to Britten
My peregrinations through the literature over the past 20 years have turned up no prior attempt to clarify systematically how cellular organization arises. However, an instructive parallel exists in Roy Britten's recent reflections on the logic underlying current models of animal development (17, 18). According to Britten (17), “Control of development is by means of local interactions rather than global control mechanisms.” As Stent had done much earlier, Britten maintains that there is no program of development, no identifiable plan. Embryos form as the result of numerous interactions among molecules and/or cells, with specific association and binding as the primary mechanism. “An organism will assemble automatically from parts (macromolecules, structures and cells) specified by nuclear control factors … Without global control systems, information for form is in the genes for structural proteins, adhesion molecules, control factors, signaling molecules, and their control regions” (17). Britten explicitly makes allowance for spatially extended influences, such as morphogen gradients and hormones, which are not precisely local but also do not contain global information specifying the form of the embryo. “Self-assembly is the logical replacement for potential overarching regulatory concepts,” for which no specific mechanisms can presently be identified (17). He acknowledges that “a cell is always required” (17) as a foundation upon which the molecules that convey developmental instructions are arrayed; but the genesis of those cells falls beyond his scope.
It is precisely that cellular substratum that concerns us in the present essay. The views put forward here mesh with Britten's in many respects but differ markedly with regard to the genesis of long-range order. Britten underscores molecules and local events; the genes that specify those molecular structures collectively encode the information for form, but he notes drily that “As yet, decoding requires an embryo” (17). The present argument does not minimize the importance of molecular associations. But it highlights the contribution of organizing principles that operate on the cellular scale, including spatial markers, vectorial physiology, gradient fields, and physical forces. And the argument leads to the conclusion that the cell represents a fundamental unit of biological organization, a state of matter that is not wholly reducible to its molecular constituents. Granted that every biological activity has a molecular basis and stems from more or less local molecular interactions, it is quite clear that the organization of a cell as a whole can only be produced by the agency of a previous cell. As I read the literature, what we have learned points plainly in an unconventional direction, one that puts cells, rather than molecules and genes, in the center of the stage.
Are Cells Computable?
To see what is meant by putting the cell in the center, let us apply to cells the question that Wolpert (166) famously posed about embryos: will the cell be “computable”? Given all the information encoded in the genome of a bacterial cell, should it be possible in principle to predict its morphology, development, and cytoplasmic organization? If the answer is no, what additional information is required? It goes without saying that such calculations are not feasible at present. Recall that one cannot yet reliably predict the folding of a protein from its amino acid sequence (although there has been steady progress towards that goal). “Computing a cell” is a trendy metaphor for understanding the logic of its development, but there is nothing whatever wrong with metaphors. Even under the best of circumstances, computing a cell of E. coli would be a crushing task if it called for tracking the movements of 50 million molecules at once; but this is not the way cells go about their business. As Wolpert (166) noted, the computational hurdles can be greatly diminished by choosing a level of descriptive complexity that is sufficiently detailed to account for the overall course of events without attending to individual molecules. Now physiology rides to the rescue, making it practicable to literally compute some cells even in our own day.
I have been much impressed by calculations and simulations that allow one to predict, in selected cases, features of spatial organization and global morphology from explicit and realistic premises. Examples include the shapes of bacterial cell poles and of whole streptococcal cells (89); apical regeneration in Acetabularia acetabulum (16, 51); growth and form in fungal hyphae (11, 47); and the self-organization of microtubules and actin meshworks (103, 104, 118). To be sure, generation of a plausible shape or pattern does not necessarily validate the premises that underlie the calculations, and more than one algorithm can generate a given form. All the same, an algorithm that correctly predicts a biological form from physiological principles deserves to be taken seriously. At the very least, these achievements suggest that, if the reductionist agenda is to be productively applied to living systems, computation must begin from physiological processes rather than molecules or genes.
So far, so good; but the prospect that cellular forms and organization can (at least in principle) be derived from cell physiology begs a deeper question. Is the choice of the physiological level just a matter of heuristic convenience, a procedure that reduces the tasks of computation and prediction to manageable dimensions, or does it correspond in some fundamental way to biological realities? Stated thus, the question becomes a version of the perennial issue of the “reducibility” of biology: can the architecture and form of cells be wholly explained in terms of the chemistry and physics of their constituent molecules? Alternatively, must any such explanation be grounded in the organized state of those molecules? Among philosophers of biology, this is a lively issue that has recently bubbled up again with reference to the spectacular advances in the molecular basis of embryonic development. For Rosenberg (138), those discoveries imply that, at least in principle, even a biological object as complex as an eye finds explanation in strictly molecular terms. For Laublicher and Wagner (93), and with qualifications also for Frost-Arnold (45), reduction fails unless the chemical capacities of molecules are located within a functional, spatially organized context.
Let us reason this out with reference to cells rather than embryos. There is agreement that cells are made of molecules and only of molecules and that everything cells are and do, including form and spatial organization, grows out of interactions among molecules. Many scientists apparently infer from these premises that cells can ultimately be fully explained by (even predicted by and computed from) the chemical characteristics of those molecules: sequences, structures, and couplings. In my judgment, what we have learned of cell growth and morphogenesis shows this hypothesis to be overly simple. Many cellular features and functions, including growth, division, and morphogenesis, are fundamentally dependent on processes that have location and direction in cell space. Moreover, these spatial parameters are propagated by mechanisms that are, at least in part, independent of molecular chemistry. The latter point is crucial, for it asserts that the spatial relationships within the cellular system are not wholly a consequence of scalar chemical properties but represent an autonomous level that wields explanatory force. I take the data to mean that the biology of cells cannot be reduced to (explained by, or computed from) molecular chemistry alone; therefore, a fully materialistic accounting must be based on molecules that form part of a spatially organized system.
To cement the point, consider an example that happens to correspond precisely to the hypothetical case dissected by Frost-Arnold (45), in his essay on what is required to refute the claim of reducibility. The zygotes of the brown algae Fucus and Silvetia spp. (previously known as Pelvetia) are, to judge by their appearance and behavior, spherically symmetrical. The axis of the developing embryo is established when the zygote germinates, at a locus determined by the direction of incident light. Zygotes will germinate even in the dark, at a locus that corresponds to the site of sperm entry (54, 92). When all the data are in, will these early stages of morphogenesis be wholly comprehensible in terms of the chemistry of the participating molecules? Obviously not; but they should be comprehensible as a spatially organized system of molecules that can take directions from environmental vectors.
Extension of this conclusion to E. coli, whose morphogenesis requires no external cues, hinges on the degree to which the propagation of spatial order can be separated from molecular chemistry. The recent elucidation of the mechanisms by which E. coli localizes the septum that will define two new cell poles illustrates the interplay of chemical and cellular factors in the genesis of spatial order. To me, it seems absurd to deny that, in this instance as in the preceding one, spatial organization is essential to a satisfying explanation of the cell. Whether or not one considers this a successful reduction to the molecular level depends on one's sense of what “molecular” means; but it had better refer to an organized system, not to chemistry alone. To rephrase this point in terms of a question posed in the introduction to this essay: dissociation of a cell into its molecular constituents does, indeed, destroy something irretrievable—the spatial organization that makes that cell alive.
And Therefore, What?
Which research programs, if any, will be affected by whether cells can ultimately be understood (“computed”) as aggregates of gene products or only as spatially organized systems? Working scientists wrestling with the intricacies of cell growth and development have become fully conscious of spatial organization. The great majority, sharply focused on cellular mechanics and their genetic basis, will take little notice of this abstract issue and continue on their way. But scholars of a more philosophical bent should find advantage in recognizing that in unicellular organisms, the relationship between molecules and life is direct and out in the open; bacteria are simpler and more tractable subjects for reflection than flies and frogs.
Even at this stage, our burgeoning knowledge of how microbes grow and make themselves raises grave doubts about the widespread conception of organisms as mere creatures of their genomes. No one has articulated that position more forcefully than Richard Dawkins (30, 31, 32): organisms are “robots” programmed by their DNA, “survival machines” that the genes construct and ride in pursuit of their sole objective, which is to reproduce themselves. Genes replicate, and in that fashion persist; bodies are disposable. A frame of reference that puts genes central is often highly illuminating, particularly when the questions turn on higher organisms and their transformation on the evolutionary time scale. However, as Dawkins himself readily acknowledges (30), the metaphor of the “selfish gene” becomes more ambiguous when applied to physiology and even heredity in asexual organisms. After all, genes can only exercise their informational powers in the context of an existing cell. Conversely, although cell continuity makes the cell itself a replicator of sorts, cells can only perpetuate themselves through the instructions encoded in their genes. The relationship between genes and cells is one of complementarity, not precedence of one over the other. If the goal is to comprehend organisms, spatial organization can never be set aside. An experimental test of this position may grow out of ongoing efforts to synthesize cells in the laboratory.
The subject most urgently in need of an infusion of spatial thinking is cell evolution, which is at last emerging as the proper focus for inquiry into the origin of life (164, 165). The problem of life's inception has been particularly attractive to chemists, who tend to equate it with the genesis of the informational macromolecules, proteins and nucleic acids. The hypothesis that life began with naked protogenes that replicated themselves in a prebiotic soup and then “learned” to encode proteins that promoted the multiplication of those protogenes has been formulated in many versions (36, 37, 79, 97, 109, 129, 130). It has sparked an intense research effort, with the synthesis of a self-replicating molecule as its holy grail; and it probably retains the loyalty of a majority of molecular scientists. But “genes first” makes no contact with biological organization and offers little insight into the origin of cells as we know them.
The proposition defended here, that the living state inherently requires a considerable degree of spatial organization, points in quite another direction. It suggests that life will have begun not with naked protogenes, but with chemical systems that progressively came to display the hallmarks of cells: boundaries, metabolism, spatial order, energy transduction, autopoiesis (60, 99, 163). Heredity based on nucleic acids will not have been first on stage, but its advent made possible evolution by variation and selection and marked the appearance of recognizable life.
Several hypotheses grounded in this viewpoint can be found in the literature and are beginning to draw attention. For Cavalier-Smith (23), life begins with protocells which already displayed that characteristic collaboration of membranes, replicators, and catalysts. Key evolutionary advances, including the origin of energy transduction, protein synthesis, and the genetic code, occurred on the outer surface of “obcells,” inside-out membranous bodies that subsequently folded to generate the topology of modern cells. Russell and his colleagues (108, 141) find the cradle of life in the interstices of alkaline seafloor seeps. They invoke the precipitation of an inorganic barrier, made of iron sulfides, that separates alkaline vent fluid from the acidic ocean water. The electrochemical potential across that primordial chemiosmotic membrane could serve as the immediate energy donor for a diversity of prebiotic processes, including eventually the replacement of inorganic reactions by organic ones, and then the emergence of free-living cells.
I rather suspect that neither of these proposals will fire the imagination of the scientific community as the quest for the self-replicating protogene clearly has. Modern science has a strong bias towards reductionist ideas, and for good reason; it is very much more difficult to formulate equally seductive notions in a synthetic, integrative mode. We urgently need a plausible and experimentally fertile hypothesis that starts with a driving force (sunlight or geochemical reactions) and builds up organic complexity in a membrane-bound, structured setting. No satisfying hypothesis of this kind is presently on the books, and in its absence holistic explorers of deep time have been unable to initiate a research tradition that can thrive in today's intellectual and fiscal climate. But I have no doubt that this is the way to go; for only through the emergence of spatially organized systems can molecules come to life.
ADDENDUM IN PROOF
Since this article was accepted for publication, Karsenti and his colleagues have demonstrated that mitotic chromosomes generate a gradient of the phosphorylated protein Ran-GTP, which alters microtubule dynamics and helps organize the mitotic spindle (M. Caudron, G. Bunt, P. Bastiaans, and E. Karsenti, Science 309:1373-1376, 2005; see also P. R. Clarke, Science 309:1334-1335, 2005). This makes a second well-documented example of the use of spatial gradients to supply positional information within the cytoplasm.
Acknowledgments
I am twice indebted to Seymour S. Cohen: for teaching me by example, nearly half a century ago, how a mature scientist handles a brash but bright young critic; and also for allowing me to requote André Gide from his book Introduction to Polyamines (Prentice-Hall, 1971). Joseph Frankel and Byron Johnson drew my attention to references that I had missed. Dennis Bray, Ruth Harold, Nick Lane, Carl Woese, and two anonymous reviewers made pointed suggestions that helped to clarify my thesis and sharpen its expression; David Ehlert drew the illustrations; and Katharine Norris prepared the manuscript for publication. Thank you all!
REFERENCES
- 1.Adams, I. R., and J. V. Kilmartin. 2000. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10:329-334. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht-Buehler, G. 1990. In defense of “non-molecular” cell biology. Internat. Rev. Cytol. 120:191-241. [DOI] [PubMed] [Google Scholar]
- 3.Arnone, M., and E. H. Davidson. 1997. The hardwiring of development: organization and function of genomic regulatory systems. Development 124:1851-1864. [DOI] [PubMed] [Google Scholar]
- 4.Ausmees, N., and C. Jacobs-Wagner. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu. Rev. Microbiol. 57:225-247. [DOI] [PubMed] [Google Scholar]
- 5.Ausmees, N., J. R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705-713. [DOI] [PubMed] [Google Scholar]
- 6.Baba, M., N. Baba, Y. Ohsumi, K. Kanaya, and M. Osumi. 1989. Three-dimensional analysis of morphogenesis induced by mating pheromone α-factor in Saccharomyces cerevisiae. J. Cell Sci. 94:207-216. [DOI] [PubMed] [Google Scholar]
- 7.Ball, P. 1999. The self-made tapestry. Oxford University Press, New York, N.Y.
- 8.Bartnicki-Garcia, S. 2002. Hyphal tip growth: outstanding questions, p. 29-58. In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, New York, N.Y.
- 9.Bartnicki-Garcia, S., D. D. Bartnicki, and G. Gierz. 1995. Determinants of fungal cell wall morphology: the vesicle supply center. Can. J. Bot. 73(Suppl. 1):S372-S378. [Google Scholar]
- 10.Bartnicki-Garcia, S., C. E. Bracker, G. Gierz, R. Lopez-Franco, and H. Lu. 2000. Mapping the growth of fungal hyphae: orthogonal cell wall expansion during tip growth and the role of turgor. Biophys. J. 79:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartnicki-Garcia, S., F. Hergert, and G. Gierz. 1989. Computer simulation of fungal morphogenesis and the mathematical basis of hyphal tip growth. Protoplasma 153:46-57. [Google Scholar]
- 12.Beisson, J., and M. Wright. 2003. Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 15:96-104. [DOI] [PubMed] [Google Scholar]
- 13.Bhalerao, R. P., and M. J. Bennett. 2003. The case for morphogens in plants. Nat. Cell Biol. 5:939-943. [DOI] [PubMed] [Google Scholar]
- 14.Breier, A., and N. R. Cozzarelli. 2004. Linear ordering and dynamic segregation of the bacterial chromosome. Proc. Natl. Acad. Sci. USA 101:9175-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bretscher, A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors and receptors. J. Cell. Biol. 160:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brière, C., and B. C. Goodwin. 1988. Geometry and dynamics of tip morphogenesis in Acetabularia. J. Theor. Biol. 131:461-475. [Google Scholar]
- 17.Britten, R. J. 1998. Underlying assumptions of developmental models. Proc. Natl. Acad. Sci. USA 95:9372-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Britten, R. J. 2003. Only details determine, p. 75-81. In G. B. Müller and S. A. Newman (ed.), Origination of organismal form: beyond the gene in developmental and evolutionary biology. MIT Press, Cambridge, Mass.
- 19.Brun, Y. V., G. Marczynski, and L. Shapiro. 1994. The expression of asymmetry during Caulobacter cell differentiation. Annu. Rev. Biochem. 63:419-450. [DOI] [PubMed] [Google Scholar]
- 20.Brunner, D., and P. Nurse. 2000. New concepts in fission yeast morphogenesis. Phil. Trans. R. Soc. Lond. B 355:873-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camazine, S., J.-L. Deneubourg, N. R. Franks, J. Sneyd, G. Theraulaz, and E. Bonabeau. 2001. Self-organization in biological systems. Princeton University Press, Princeton, N.J.
- 22.Cavalier-Smith, T. 2000. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5:174-182. [DOI] [PubMed] [Google Scholar]
- 23.Cavalier-Smith, T. 2001. Obcells as proto-organisms: membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells and photosynthesis. J. Mol. Evol. 53:555-595. [DOI] [PubMed] [Google Scholar]
- 24.Chang, F. 2001. Establishment of a cellular axis in fission yeast. Trends Genet. 17:273-278. [DOI] [PubMed] [Google Scholar]
- 25.Chang, F., and M. Peter. 2003. Yeasts make their mark. Nat. Cell Biol. 5:294-299. [DOI] [PubMed] [Google Scholar]
- 26.Chant, J. 1999. Polarity in yeast. Annu. Rev. Cell Dev. Biol. 15:365-391. [DOI] [PubMed] [Google Scholar]
- 27.Cosgrove, D. J. 2000. Loosening of plant cell walls by expansins. Nature 407:321-326. [DOI] [PubMed] [Google Scholar]
- 28.Danchin, A., P. Guerdoux-Jamet, I. Moszer, and P. Nitschkè. 2000. Mapping the bacterial cell architecture into the chromosome. Phil. Trans. R. Soc. Lond. B 355:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 30.Dawkins, R. 1982. The extended phenotype, p. 176-177. Oxford University Press, Oxford, United Kingdom.
- 31.Dawkins, R. 1989. The selfish gene, new edition. Oxford University Press, Oxford, United Kingdom.
- 32.Dawkins, R. 1995. River out of Eden. Harper Collins Publishers, New York, N.Y.
- 33.Drees, B. L., B. Sundin, E. Brazeau, J. L. Cavistone, G.-C. Chen, W. Guo, K. G. Kozminsky, M. W. Lau, J. J. Moskow, A. Tong, L. R. Schenkman, A. McKenzie III, P. Brennwald, M. Longtine, E. Bi, C. Chan, P. Novick, C. Boone, J. R. Pringle, T. M. Davis, S. Fields, and D. G. Drubin. 2001. A protein interaction map for cell polarity development. J. Cell. Biol. 154:549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drubin, D. G. (ed.). 2000. Cell polarity. Oxford University Press, New York, N.Y.
- 35.Du, Y., S. Ferro-Novick, and P. Novick. 2004. Dynamics and inheritance of the endoplasmic reticulum. J. Cell Sci. 117:2871-2878. [DOI] [PubMed] [Google Scholar]
- 36.Eigen, M. 1992. Steps towards life: a perspective on evolution. Oxford University Press, New York, N.Y.
- 37.Elitzur, A. C. 1994. Let there be life: thermodynamic reflections on biogenesis and evolution. J. Theor. Biol. 168:429-459. [DOI] [PubMed] [Google Scholar]
- 38.England, J. C., and J. W. Gober. 2001. Cell cycle control of cell morphogenesis in Caulobacter. Curr. Opin. Microbiol. 4:674-680. [DOI] [PubMed] [Google Scholar]
- 39.Errington, J. 2003. The bacterial actin cytoskeleton. ASM News 69:608-614. [Google Scholar]
- 40.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feijo', J. A., J. Sainhas, T. Holdaway-Clarke, M. Sofia Cordeiro, J. G. Kunkel, and P. K. Hepler. 2001. Cellular oscillations and the regulation of growth: the pollen tube paradigm. BioEssays 23:86-94. [DOI] [PubMed] [Google Scholar]
- 42.Frankel, J. 1989. Pattern formation: ciliate studies and models. Oxford University Press, New York, N.Y.
- 43.Frankel, J. 1992. Positional information in cells and organisms. Trends Cell Biol. 1:256-260. [DOI] [PubMed] [Google Scholar]
- 44.Frankel, J. 2000. Cell polarity in ciliates, p. 78-105. In D. G. Drubin (ed.), Cell polarity. Oxford University Press, New York, N.Y.
- 45.Frost-Arnold, G. 2004. How to be an anti-reductionist about developmental biology: response to Laublicher and Wagner. Biol. Phil. 19:75-91. [Google Scholar]
- 46.Gerdes, K., J. Møller-Jensen, G. Ebersbach, T. Kruse, and K. Nordström. 2004. Bacterial mitotic machineries. Cell 116:359-366. [DOI] [PubMed] [Google Scholar]
- 47.Gierz, G., and S. Bartnicki-Garcia. 2001. A three-dimensional model of fungal morphogenesis based on the vesicle supply center concept. J. Theor. Biol. 208:151-164. [DOI] [PubMed] [Google Scholar]
- 48.Gilbert, S. F., J. M. Opitz, and R. A. Raff. 1996. Resynthesizing evolutionary and developmental biolog. Dev. Biol. 173:357-372. [DOI] [PubMed] [Google Scholar]
- 49.Gitai, Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120:577-586. [DOI] [PubMed] [Google Scholar]
- 50.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodwin, B. C. 1994. How the leopard changed its spots: the evolution of complexity. Simon and Schuster, New York, N.Y.
- 52.Green, P. B. 1987. Inheritance of pattern: analysis from phenotype to gene. Am. Zool. 27:657-673. [Google Scholar]
- 53.Gurdon, J. B., and P.-Y. Bourillot. 2001. Morphogen gradient interpretation. Nature 413:797-803. [DOI] [PubMed] [Google Scholar]
- 54.Hable, W. E., N. R. Miller, and D. L. Kropf. 2003. Polarity establishment requires dynamic actin in fucoid zygotes. Protoplasma 221:193-204. [DOI] [PubMed] [Google Scholar]
- 55.Hagan, I. M. 1998. The fission yeast microtubule cytoskeleton. J. Cell Sci. 111:1603-1612. [DOI] [PubMed] [Google Scholar]
- 56.Hale, C., H. Meinhardt, and P. J. A. de Boer. 2001. Dynamic localization of the cell division regulator MinE in Escherichi coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harold, F. M. 1990. To shape a cell: an inquiry into the causes of morphogenesis of microorganisms. Microbiol. Rev. 54:381-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harold, F. M. 1995. From morphogenes to morphogenesis. Microbiology 141:2765-2778. [DOI] [PubMed] [Google Scholar]
- 59.Harold, F. M. 1997. How hyphae grow: morphogenesis explained? Protoplasma 197:137-147. [Google Scholar]
- 60.Harold, F. M. 2001. The way of the cell: molecules, organisms and the order of life. Oxford University Press, New York, N.Y.
- 61.Harold, F. M. 2001. Postscript to Schrödinger: so what is life? ASM News 67:611-616. [Google Scholar]
- 62.Harold, F. M. 2002. Force and compliance: rethinking morphogenesis in walled cells. Fungal Genet. Biol. 37:271-282. [DOI] [PubMed] [Google Scholar]
- 63.Harold, R. L., N. P. Money, and F. M. Harold. 1996. Growth and morphogenesis in Saprolegnia ferax: is turgor required? Protoplasma 191:105-114. [Google Scholar]
- 64.Harris, S. D., and M. Momany. 2001. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 65.Hayles, J., and P. Nurse. 2001. A journey into space. Nat. Rev. Mol. Cell. Biol. 2:647-656. [DOI] [PubMed] [Google Scholar]
- 66.Heath, I. B. (ed.). 1990. Tip growth in plant and fungal cells. Academic Press, San Diego, Calif.
- 67.Heath, I. B. 1994. The cytoskeleton in hyphal growth, organelle movements, and mitosis, p. 43-65. In J. G. H. Wessels and E. Meinhardt (ed.), The mycota, vol. I: growth, differentiation and sexuality. Springer Verlag, Berlin, Germany.
- 68.Holdaway-Clarke, T. L., and P. K. Hepler. 2003. Control of pollen tube growth: role of ion gradients and fluxes. New Phytol. 159:539-563. [DOI] [PubMed] [Google Scholar]
- 69.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howard, J., and A. A. Hyman. 2003. Dynamics and mechanics of the microtubule plus end. Nature 422:753-758. [DOI] [PubMed] [Google Scholar]
- 71.Hyman, A. A., and E. Karsenti. 1996. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell 84:401-410. [DOI] [PubMed] [Google Scholar]
- 72.Iftode, F., and A. Fleury-Aubusson. 2003. Structural inheritance in Paramecium: ultrastructural evidence for basal body and associated rootlets polarity transmission through binary fission. Biol. Cell 95:39-51. [DOI] [PubMed] [Google Scholar]
- 73.Ingber, D. E. 2003a. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell Sci. 116:1157-1173. [DOI] [PubMed] [Google Scholar]
- 74.Ingber, D. E. 2003b. Tensegrity II. How structural networks influence cellular information processing networks. J. Cell Sci. 116:1397-1408. [DOI] [PubMed] [Google Scholar]
- 75.Jacob, F. 1973. The logic of life: a history of heredity. Pantheon Books, New York, N.Y.
- 76.Jaffe, L. F. 1968. Localization in the developing Fucus egg and the general role of localizing currents. Adv. Morphogen. 7:295-328. [DOI] [PubMed] [Google Scholar]
- 77.Jesch, S. A. 2002. Inheriting a structural scaffold for Golgi biosynthesis. BioEssays 24:584-587. [DOI] [PubMed] [Google Scholar]
- 78.Jones, L. F., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 79.Joyce, G. F. 2002. The antiquity of RNA-based evolution. Nature 418:214-221. [DOI] [PubMed] [Google Scholar]
- 80.Judd, E. M., K. R. Ryan, W. R. Moemer, L. Shapiro, and H. H. McAdams. 2003. Fluorescence bleaching reveals compartment formation prior to cell division in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 100:8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karsenti, E., and I. Vernos. 2001. The mitotic spindle: a self-made machine. Science 294:543-547. [DOI] [PubMed] [Google Scholar]
- 82.Katz, M. 1986. Templets and the explanation of complex patterns. Cambridge University Press, Cambridge, United Kingdom.
- 83.Kessler, M. A., and B. T. Werner. 2003. Self-organization of sorted patterned ground. Science 299:380-384. [DOI] [PubMed] [Google Scholar]
- 84.Kim, G. H., T. A. Klotchkova, and Y.-M. Kang. 2001. Life without a cell membrane: regeneration of protoplasts from disintegrated cells of the marine green alga Bryopsis plumosa. J. Cell. Sci. 114:2009-2014. [DOI] [PubMed] [Google Scholar]
- 85.Kirschner, M., and T. Mitchison. 1986. Beyond self-assembly: from microtubules to morphogenesis. Cell 45:329-342. [DOI] [PubMed] [Google Scholar]
- 86.Kirschner, M., J. Gerhart, and T. Mitchison. 2000. Molecular “vitalism.” Cell 100:79-88. [DOI] [PubMed] [Google Scholar]
- 87.Koch, A. L. 1988. Biophysics of the bacterial wall viewed as a stress-bearing fabric. Microbiol. Rev. 52:337-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koch, A. L. 1994. The problem of hyphal growth in streptomycetes and fungi. J. Theor. Biol. 171:137-150. [Google Scholar]
- 89.Koch, A. L. 1995. Bacterial growth and form. Chapman and Hall, New York, N.Y.
- 90.Koch, A. L. 2000. The bacterium's way for safe enlargement and division. Appl. Environ. Microbiol. 66:3657-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koch, A., M. L. Higgins, and R. J. Doyle. 1981. Surface tension-like forces determine bacterial shapes: Streptococcus faecium. J. Gen. Microbiol. 123:151-161. [DOI] [PubMed] [Google Scholar]
- 92.Kropf, D. L. 1994. Cytoskeletal control of cell polarity in a plant zygote. Dev. Biol. 165:361-371. [DOI] [PubMed] [Google Scholar]
- 93.Laublicher, M. D., and G. P. Wagner. 2001. How molecular is molecular developmental biology? A reply to Alex Rosenberg's reductionism redux: computing the embryo. Biol. Phil. 16:53-68. [Google Scholar]
- 94.Lawrence, P. A. 2001. Morphogens: how big is the big picture? Nat. Cell Biol. 31:E151-E154. [DOI] [PubMed] [Google Scholar]
- 95.Lehn, J.-M. 2002. Toward self-organization and complex matter. Science 295:2400-2403. [DOI] [PubMed] [Google Scholar]
- 96.Lew, R. R., N. N. Levina, S. K. Walker, and A. Garrill. 2004. Turgor regulation in hyphal organisms. Fungal Genet. Biol. 41:1007-1015. [DOI] [PubMed] [Google Scholar]
- 97.Lifson, F. 1997. On the crucial stages in the origin of animate matter. J. Mol. Evol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 98.Luby-Phelps, K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192:189-221. [DOI] [PubMed] [Google Scholar]
- 99.Luisi, P. L. 2003. Autopoiesis: a review and a reappraisal. Naturwissenschaften 90:49-59. [DOI] [PubMed] [Google Scholar]
- 100.Lutkenhaus, J., and S. G. Adinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 101.Lybarger, S., and J. R. Maddock. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Madden, K., and M. Snyder. 1998. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52:687-744. [DOI] [PubMed] [Google Scholar]
- 103.Maly, I. V., and G. Borisy. 2001. Self-organization of a propulsive actin network as an evolutionary process. Proc. Natl. Acad. Sci. USA 98:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maly, I. V., and G. Borisy. 2002. Self-organization of treadmilling microtubules into a polar array. Trends Cell Biol. 12:462-465. [DOI] [PubMed] [Google Scholar]
- 105.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 106.Marks, J., and J. S. Hyams. 1985. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur. J. Cell Biol. 39:27-32. [Google Scholar]
- 107.Marshall, E. 2002. Venter gets down to life's basics. Science 298:1701. [DOI] [PubMed] [Google Scholar]
- 108.Martin, W., and M. J. Russell. 2003. On the origin of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil. Trans. R. Soc. Lond. B 358:59-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maynard Smith, J., and E. Szathmary. 1995. The major transitions of evolution. W. H. Freeman, New York, N.Y.
- 110.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 111.McNeil, P. L., and R. A. Steinhardt. 2003. Plasma membrane repair and disruption. Annu. Rev. Cell Biol. 19:697-731. [DOI] [PubMed] [Google Scholar]
- 112.Meinhardt, H., and P. A. J. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillation of Min proteins, and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Misteli, T. 2001. The concept of self-organization in cellular architecture. J. Cell Biol. 155:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Money, N. P. 1997. Wishful thinking of turgor revisited: the mechanics of fungal growth. Fungal Genet. Biol. 21:173-187. [Google Scholar]
- 115.Munro, S. 2002. More than one way to replicate the Golgi apparatus. Nat. Cell Biol. 4:E223-E224. [DOI] [PubMed] [Google Scholar]
- 116.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nanninga, N. 2001. Cytokinesis in prokaryotes and eukaryotes: common principles and different solutions. Microbiol. Mol. Biol. Rev. 65:319-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nèdèlec, F. J., T. Surrey, A. C. Maggs, and S. Leibler. 1997. Self-organization of microtubules and motors. Nature 389:305-308. [DOI] [PubMed] [Google Scholar]
- 119.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. L. Reznikoff, M. Riley, and H. E. Umbarger (ed.). Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 120.Nelson, W. J. 2003. Adaptation of core mechanisms to generate cell polarity. Nature 422:766-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nijhout, H. F. 1990. Metaphors and the role of genes in development. BioEssays 12:441-446. [DOI] [PubMed] [Google Scholar]
- 122.Noda, T., K. Suzuki, and Y. Ohsumi. 2002. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12:231-235. [DOI] [PubMed] [Google Scholar]
- 123.Onoda, T., A. Oshima, S. Nakano, and A. Matsuno. 1987. Morphology, growth and reversion in a stable L-form of Escherichia coli K12. J. Gen. Microbiol. 133:527-534. [DOI] [PubMed] [Google Scholar]
- 124.Pantaloni, D., C. Le Clainche, and M.-F. Carlier. 2001. Mechanism of actin-based motility. Science 292:1502-1506. [DOI] [PubMed] [Google Scholar]
- 125.Penman, S. 1995. Rethinking cell structure. Proc. Natl. Acad. Sci. USA 92:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell. Sci. 111:867-876. [DOI] [PubMed] [Google Scholar]
- 127.Pollack, G. H. 2001. Cells, gels and the engines of life: a new, unifying approach to cell function. Ebner and Sons, Seattle, Wash.
- 128.Pringle, J. R., E. Bi, H. A. Harkins, J. E. Zahner, C. D. Virgilio, J. Chant, K. Corrado, and H. Farres. 1995. Establishment of cell polarity in yeast. Cold Spring Harbor Symp. Quant. Biol. 60:729-744. [DOI] [PubMed] [Google Scholar]
- 129.Pross, A. 2003. The driving force for life's emergence: kinetic and thermodynamic considerations. J. Theor. Biol. 220:393-406. [DOI] [PubMed] [Google Scholar]
- 130.Pross, A. 2004. Causation and the origin of life: metabolism or replication first? Orig. Life Evol. Biosph. 25:297-334. [DOI] [PubMed] [Google Scholar]
- 131.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365-375. [DOI] [PubMed] [Google Scholar]
- 132.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell. Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 133.Rafelski, S. M., and J. A. Theriot. 2004. Crawling toward a unified model of cell motility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 73:209-239. [DOI] [PubMed] [Google Scholar]
- 134.Rayle, D. L., and R. E. Cleland. 1992. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99:1271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Riquelme, M., G. Gierz, and S. Bartnicki-Garcia. 2000. Dynein and dynactin deficiencies affect the formation and function of the Spitzenkörper and distort hyphal morphogenesis of Neurospora crassa. Microbiology 146:1743-1752. [DOI] [PubMed] [Google Scholar]
- 136.Riquelme, M., C. R. Reynaga-Peña, G. Gierz, and S. Bartnicki-Garcia. 1998. What determines growth direction in fungal hyphae? Fungal Genet. Biol. 24:101-109. [DOI] [PubMed] [Google Scholar]
- 137.Robinson, K. R., M. Wozniak, R. Pu, and M. Messerli. 1999. Symmetry breaking in the zygotes of the fucoid algae: controversies and recent progress. Curr. Top. Dev. Biol. 44:101-125. [DOI] [PubMed] [Google Scholar]
- 138.Rosenberg, A. 1997. Reductionism redux: computing the embryo. Biol. Philos. 12:445-470. [Google Scholar]
- 139.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-428. [DOI] [PubMed] [Google Scholar]
- 140.Rothfield, L., Y.-L. Shih, and G. King. 2001. Polar explorers: membrane proteins that determine division site placement. Cell 106:13-16. [DOI] [PubMed] [Google Scholar]
- 141.Russell, M. J., and A. J. Hall. 1997. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J. Geol. Soc. Lond. 154:377-402. [DOI] [PubMed] [Google Scholar]
- 142.Ryan, K. R., S. Huntwork, and L. Shapiro. 2004. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl. Acad. Sci. USA 101:7415-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sawin, K. E., and H. A. Snaith. 2004. Role of microtubules and Tea1p in establishment and maintenance of fission yeast cell polarity. J. Cell Sci. 117:689-700. [DOI] [PubMed] [Google Scholar]
- 144.Schliwa, M. 2002. The evolving complexity of cytoplasmic structure. Nat. Rev. Mol. Biol. 3:291-296. [DOI] [PubMed] [Google Scholar]
- 145.Seemann, J., E. Jokitalo, M. Pypaert, and G. Warren. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407:1022-1026. [DOI] [PubMed] [Google Scholar]
- 146.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89-98. [DOI] [PubMed] [Google Scholar]
- 147.Shapiro, L., H. H. McAdams, and A. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 148.Shih, Y.-L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Silverman-Gavrila, L. B., and R. R. Lew. 2001. Regulation of the tip-high [Ca2+] gradient in growing hyphae of the fungus Neurospora crassa. Eur. J. Cell. Biol. 80:379-390. [DOI] [PubMed] [Google Scholar]
- 150.Silverman-Gavrila, L. B., and R. R. Lew. 2003. Calcium gradient dependence of Neurospora crassa hyphal growth. Microbiology 149:2475-2485. [DOI] [PubMed] [Google Scholar]
- 151.Strohman, R. C. 1997. The coming Kuhnian revolution in biology. Nat. Biotechnol. 15:194-200. [DOI] [PubMed] [Google Scholar]
- 152.Surrey, T., F. Nèdèlec, S. Leibler, and E. Karsenti. 2001. Physical properties determining self-organization of motors and microtubules. Science 292:1167-1172. [DOI] [PubMed] [Google Scholar]
- 153.TerBush, D. R., T. Maurice, D. Roth, and P. Novick. 1999. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15:6494-6583. [PMC free article] [PubMed] [Google Scholar]
- 154.Torralba, S., and I. B. Heath. 2001. Cytoskeletal and Ca2+ regulation of hyphal tip growth and initiation. Curr. Top. Dev. Biol. 51:135-187. [DOI] [PubMed] [Google Scholar]
- 155.Viollier, P. H., N. Sternheim, and L. Shapiro. 2002. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc. Natl. Acad. Sci. USA 99:13831-13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movements of individual chromosomes to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. USA 101:9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang, H. X. Tang, J. Li, S. Trautmann, D. Balasundaram, D. McCollum, and M. K. Balasubramanian. 2002. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 13:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weldlich-Soldner, R., and R. Li. 2003. Spontaneous cell polarization: undermining determinism. Nat. Cell Biol. 5:267-269. [DOI] [PubMed] [Google Scholar]
- 159.Wessels, J. G. H. 1986. Cell wall synthesis in apical hyphal growth. Int. Rev. Cytol. 104:37-79. [Google Scholar]
- 160.Wessels, J. G. H. 1993. Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 123:397-413. [DOI] [PubMed] [Google Scholar]
- 161.Whitesides, G. M., and M. Boncheva. 2002. Beyond molecules: self-assembly of mesoscopic and macroscopic components. Proc. Natl. Acad. Sci. USA 99:4769-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Whitesides, G. M., and B. Grzybowski. 2002. Self-assembly at all scales. Science 295:2418-2421. [DOI] [PubMed] [Google Scholar]
- 163.Wicken, J. S. 1987. Evolution, thermodynamics and information: extending the Darwinian Program. Oxford University Press, New York, N.Y.
- 164.Woese, C. R. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. USA 99:8742-8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Woese, C. R. 2004. A new biology for a new century. Microbiol. Mol. Biol. Rev. 68:173-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wolpert, L. 1994. Do we understand development? Science 266:571-572. [DOI] [PubMed] [Google Scholar]
- 167.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]