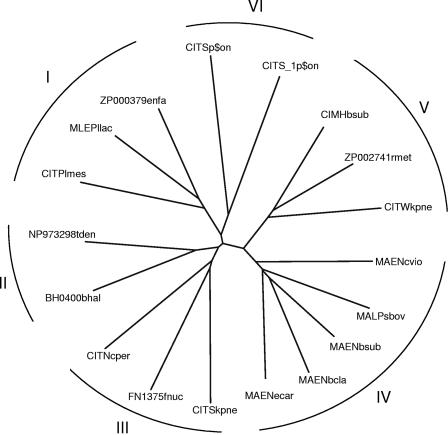
FIG. 1.
Unrooted phylogenetic tree of members of the 2HCT family. Phylogenetic relationships were analyzed with the CLUSTAL W program using the default settings (141). The tree was generated with the DRAWTREE program in the Phylip package (J. Felsenstein, PHYLIP (Phylogeny Inference Package), version 3.6.a3, Department of Genome Sciences, University of Washington, Seattle, 2002). The tree is based on the C-terminal part of the multiple-sequence alignment, which contains the fewest gaps (positions 300 to 500 in Fig. 5A). Sequences included in the alignment correspond to the “typical” sequences in the “2HCT” column of Table 1. All other members of the 2HCT transporter family share over 60% sequence identity with one of the “typical” sequences. The six clusters in the tree are indicated by I to VI.