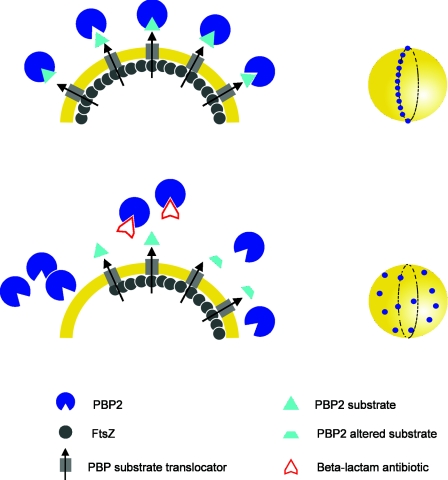
FIG. 6.
Model for PBP2 localization by substrate recognition in S. aureus. In a wild-type cell growing in the absence of inhibitors (upper panels), the substrate of PBP2 is exported from the inside of the cell at the division site, possibly because the translocator of the PBP2 substrate (presumably an FtsW homologue) localizes at the division site by interaction with the divisome. PBP2 recognizes and binds the substrate, therefore localizing at the division site, initially forming a ring around the cell (cell on the upper right). Several events can lead to delocalization of PBP2 (lower panels): depletion of FtsZ leads to delocalization of the substrate translocator and therefore to a nonlocalized export of the PBP substrate; inactivation of the PBP2 binding site by β-lactam antibiotics or alteration or blockage of the substrate (for example, by addition of d-cycloserine or vancomycin, respectively [see text]) prevents enzyme-substrate binding. Both delocalization of the substrate and inability of PBP2 to bind its substrate can cause the loss of the precise localization of PBP2 at the division septum, and the protein becomes dispersed over the entire surface of the cell (cell on the lower right).