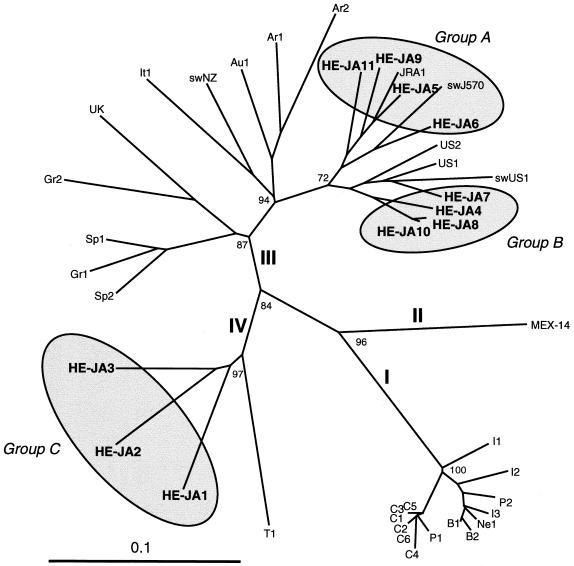
FIG. 2.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence of the ORF1 region (287 nt) of 42 human and swine HEV isolates. In addition to 21 reported human and swine HEV isolates of genotypes I to IV whose entire or nearly entire sequence is known (see Table 4 for the names of the isolates and relevant accession numbers), 10 reported isolates of genotype III whose partial sequence of 287 or 371 nt is available, as well as the 11 HEV isolates found in the present study, which are indicated in boldface type, were included for comparison. The names (accession numbers) of the 10 reported isolates are as follows: Ar1 (AF264009) and Ar2 (AF264010) from Argentina, Au1 (AF279122) from Austria, Gr1 (AF110388) and Gr2 (AF110389) from Greece, It1 (AF110387) from Italy, swNZ (AF215661) from New Zealand, Sp1 (AF195064) and Sp2 (AF195065) from Spain, and UK (AJ315768) from the United Kingdom. Genotype designations I to IV are in accordance with the recent report by Schlauder and Mushahwar (36). Bootstrap values of >70% are indicated for the major nodes as a percentage of the data obtained from 1,000 resamplings. For visual clarity, HEV isolates of Japanese origin, including those described in previous reports (JRA1 and swJ570) (25, 38), are indicated by shaded circles with the tentative designation of groups A to C (see Table 4).