Abstract
The oligomeric c-rings of Na+-translocating F1F0 ATP synthases exhibit unusual stability, resisting even boiling in SDS. Here, we show that the molecular basis for this remarkable property is intersubunit crossbridging by Na+ or Li+ ions. The heat stability of c11 was dependent on the presence of Na+ or Li+ ions. For equal stability, 10 times higher Li+ than Na+ concentrations were required, reflecting the 10 times lower binding affinity for Li+ than for Na+. In a recent structural model of c11, the Na+ or Li+ binding ligands are located on neighboring c-subunits, which thus become crossbridged by the binding of either alkali ion with a concomitant increase in the stability of the ring. Site-directed mutagenesis strengthens the essential role of glutamate 65 in the crossbridging of the subunits and also corroborates the proposed stabilizing effect of an ion bridge including aspartate 2.
Introduction
The F1F0 ATP synthase is the key enzyme for ATP synthesis in almost every living cell. The enzyme is a rotary engine consisting of an F0 portion, where torque is generated upon ion translocation, and an F1 sector, where ATP synthesis is performed. This process requires large conformational changes at the catalytic β subunits, which are elicited by the rotation of the rotor subunits (cnγε) versus the stator subunits (ab2α3β3δ) (Boyer, 1993; Abrahams et al., 1994; Noji et al., 1997). The membrane-bound F0 sector consists of subunits ab2cn, and recent structural work has shown that the number of csubunits forming the rotor ring varies among species, being 10 for yeast mitochondria (Stock et al., 1999), 11 for the bacterium Ilyobacter tartaricus (Stahlberg et al., 2001) and 14 for spinach chloroplasts (Seelert et al., 2000).
Much of our knowledge on the ion path across the membrane and the torque-generating mechanism stems from investigations of the closely related Na+-translocating F1F0 ATP synthases from Propionigenium modestum and I. tartaricus. The Na+ binding sites on c11 with ligands from Q32, E65 and S66 (Kaim et al., 1997) are freely accessible from the cytoplasmic reservoir (Kaim et al., 1998), yet they are located in the middle of the membrane (von Ballmoos et al., 2002). Hence, 11 c11 intrinsic access channels leading to these sites have been proposed, which is compatible with the structure (Vonck et al., 2002). This shows 11 closely spaced N-terminal helices forming an inner ring and 11 C-terminal outer helices packed in a staggered position to the inner helices. This leaves enough space between an inner helix and two outer helices for an ion access channel to the binding site.
It has been demonstrated that the membrane potential plays the pivotal role in the ion translocation and torque-generating mechanism (Kaim and Dimroth, 1999), and a functional model that is based on these observations has been presented (Dimroth et al., 1999). However, for a detailed understanding of the F0 motor, more structural information is required. In this study, we addressed the question of why the c-oligomer from Na+-translocating F1F0 ATP synthases has such a stable quaternary structure. We show that the stability is dependent on the presence of Na+ or Li+, which is consistent with the view that these alkali ions form intersubunit crossbridges between neighboring subunits.
Results
Effect of Na+ or Li+ on the stability of c11 from I. tartaricus
The c-ring assemblies of the three Na+-translocating ATP synthases investigated exhibit an extraordinary stability, resisting even boiling with SDS for several minutes (Laubinger and Dimroth, 1987; Neumann et al., 1998; Aufurth et al., 2000). This stability has greatly facilitated previous structural work with the c11 oligomer from I. tartaricus, but the molecular basis for it was unknown (Vonck et al., 2002). As complex formation with Na+ ions might contribute to the stability of the ring, we investigated the oligomeric status by SDS–PAGE after pretreatment of the samples at 95°C for various times and at various Na+ concentrations. The result shows that the c11 oligomer dissociates rapidly into the monomeric units if no Na+ ions were added during the heat treatment (Figure 1A). Approximately half the amount of the oligomeric complex has disappeared after 1 min at 95°C, and further dissociation was observed on prolonged heat treatments. This heat treatment not only led to the dissociation into the monomeric units, but also to increasing amounts of aggregation products. The heat stability of the c-ring increased in the presence of 0.1 mM NaCl, and at 10 mM NaCl no dissociation was apparent even after heating the sample for 60 min (Figure 1B). As Li+ is an alternative coupling ion of the I. tartaricus ATP synthase binding to the csubunits with ligands from E65 and S66 (Kaim et al., 1997), we investigated the effect of Li+ on the heat stability of c11. The results show that LiCl also protected the c11 oligomer from the dissociation by heat treatment, but for similar protection ∼10 times higher LiCl than NaCl concentrations were required (Figure 1C). These data reflect perfectly the reduced affinity of the site for Li+, as shown by a 10 times higher Km value for Li+ than for Na+ (Kluge and Dimroth, 1993a).
Figure 1.
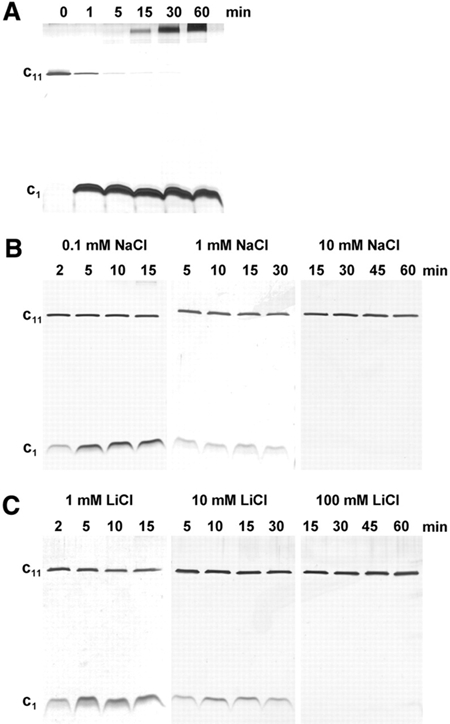
Stability of the I. tartaricus c11 oligomer. Purified c11 ring (1 μg) from I. tartaricus was incubated in buffer containing Tris–HCl pH 8.0 and 1% octylglucoside at 95°C. The incubation mixtures contained (A) no alkali ion addition, (B) NaCl and (C) LiCl in the concentrations indicated. Samples were taken after the incubation times indicated and analyzed by SDS–PAGE. The gels were stained with silver.
Effect of mutations of cE65 on the stability of c11
From the results described above, we reasoned that mutants that abolish the Na+ binding sites on c11 will destabilize the oligomeric ring. To investigate the effect of mutagenesis on the c-ring stability, we first analyzed whether c11 could be heterologously expressed in Escherichia coli. For this purpose, we switched to the very similar c11 from P. modestum, because plasmid pt7c harboring the c-subunit gene from P. modestum was already available in our laboratory (Matthey et al., 1997). The gene encoding the csubunit was expressed from this plasmid, and the c11 oligomer was purified from the recombinant E. coli cells by the same protocol as used for the isolation of c11 from I. tartaricus wild-type cells. The results of SDS–PAGE show that the c11 oligomer has been isolated from the recombinant E. coli clone and that it is of similar stability as c11 isolated from the wild type (Figure 2). Upon acidification with trichloroacetic acid, the c11 oligomer was completely dissociated into the monomeric units (Figure 2, lane 2). In parallel, recombinant E. coli cells were grown harboring plasmid pt7c with the point mutations cE65Q, cE65C or cE65D. All these strains expressed the mutant csubunit gene in amounts similar to that of the wild-type gene, as shown by the presence of the c-monomer after extraction with chloroform/methanol and analysis by SDS–PAGE. In contrast, no c-subunit could be identified by this method, with the control strain containing the empty plasmid pt7-7. However, attempts to purify c11 with either of these cE65 mutations failed. These results indicate either that the mutant c-subunits are not properly assembled into the c11 oligomer or that the c11 oligomer with either cE65 mutation is unstable and does not survive the isolation procedure. As cE65 is required for Na+ binding, this will be abolished by the mutations and therefore no stabilization of the ring by Na+ crossbridging is possible. As the isolation includes heating to 65°C for 10 min, a c-ring that is not stabilized by Na+ crossbridging is expected to dissociate (cf. Figure 1A).
Figure 2.
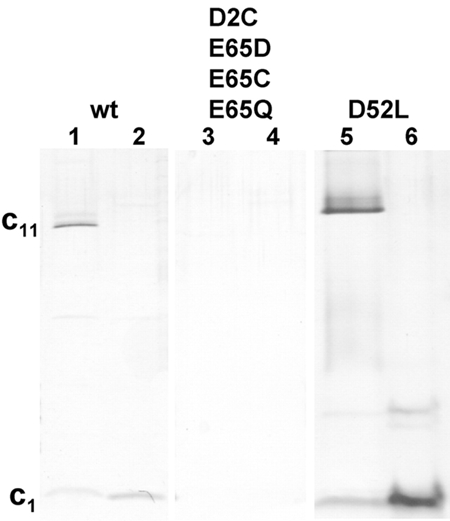
Stability of the P. modestum c11 oligomer with point mutations. Mutants were constructed in the plasmid pt7c. Mutant proteins were synthesized in E. coli BL21(DE3) harboring plasmid pt7c with the mutation and recombinant c-oligomers were purified as described in Methods. The isolated c11 rings (lanes 1, 3 and 5) were applied to SDS–PAGE gels, together with trichloroacetic acid-treated samples (lanes 2, 4 and 6) (Matthey et al., 1997). The gels were stained with silver.
Effect of mutations of cD52 or cD2 on the stability of c11
From structural work, it has been proposed that salt bridges between adjacent subunits including cD2 and cD52 of I. tartaricus (equal to P. modestum) could contribute to the stability of the c11 ring (Vonck et al., 2002). To test this hypothesis, we constructed the point mutations cD2C (Kaim et al., 2002) and cD52L. A stable c-ring was obtained from the mutant cD52L but not from the mutant cD2C (Figure 2). Therefore, aspartate 52 seems not to be required to stabilize the oligomeric structure, but aspartate 2 could play a role in the ring stability.
Effect of pH on the stability of c11
Salt bridges would also be destroyed below the pH of the dissociation constant of the acid or above the pH of the dissociation constant of the base forming the bridge. We investigated the stability of c11 at pH values ranging from 3.5 to 13. For this purpose, the isolated c11 ring was added to buffer of a certain pH containing 0.1% lauroylsarcosine and heated to 95°C for 60 min, after which the sample was analyzed by SDS–PAGE. The results summarized in Table 1 indicate that the c-ring is completely dissociated into the monomeric units during heating in the absence of Na+, confirming the results shown in Figure 1. At 3.4 mM Na+, the ring survived the heat treatment at pH values ranging from 4.5 to 11.5, but increasing the pH to 12–13 led to a significant destabilization of the ring. However, on increasing the Na+ concentration to 10 or 100 mM, part of the ring structures even survived heating at pH 12.5 or 13, respectively. The ring stability decreased rapidly below pH 4.5, and at pH 3.5 the ring dissociated completely upon heating, irrespective of the Na+ concentration added. This is to be expected, because the pK value of the Na+ binding cE65 is ∼7.0 in the absence of Na+ and decreases at elevated Na+ concentrations (Kluge and Dimroth, 1993b). At pH 3.5, however, the glutamate will be completely protonated so that Na+ is no longer able to bind.
Table 1.
Stability of I. tartaricus c11 at different pH values after 1 h of incubation at 95°C
| pH | Na+ (mM) |
|||
|---|---|---|---|---|
| 0 | 3.4 | 10 | 100 | |
| 3.5 | − | − | − | − |
| 4.0 | − | + | + | + |
| 4.5 | − | ++ | ++ | ++ |
| 8.0 | − | ++ | ++ | ++ |
| 11.5 | − | ++ | ++ | ++ |
| 12.0 | − | + | ++ | ++ |
| 12.5 | − | − | + | ++ |
| 13.0 | − | − | − | + |
++, 90–100% c11; +, 10–90% c11; −, 0–10% c11.
Discussion
The most important result of this study is the attribution of the extraordinary stability of the c11 oligomer of the Na+-translocating F1F0 ATP synthases from I. tartaricus or P. modestum to its Na+ liganded complex. In contrast, the Na+-free c11 oligomer dissociates readily into the monomeric units upon heating. These results significantly strengthen our recent structurally based proposal of the Na+ coordination sphere in which the Na+ binding site residues Q32 and S66 are contributed from the inner and outer helix, respectively, of one monomeric unit, whereas the Na+ binding E65 residue is contributed from the outer helix of the adjacent monomeric unit (Vonck et al., 2002). A model of the Na+ coordination sphere is shown in Figure 3. It clearly emphasizes the role of Na+ in bridging two adjacent subunits within the c-ring, which is obviously the rationale for its extraordinary stability. In previous mutagenesis studies, it was shown that the site also accommodates Li+ binding with E65 and S66 as ligands (Kaim et al., 1997). It was also shown that the dissociation constant for Li+ exceeds that for Na+ by a factor of 10 (Kluge and Dimroth, 1993a). Here, we observed that Na+ or Li+ at 10 times higher concentrations protected the c-ring from dissociation by heat treatment to a similar extent. Li+ ions will be liganded by S66 from the outer helix of one subunit and E65 from the outer helix of the adjacent subunit. Hence, Li+ performs crossbridging of adjacent subunits of the ring, which results in a similar stabilization to that observed with Na+, albeit at 10 times higher concentrations. Another Na+-translocating F-type ATP synthase was described in Acetobacterium woodii, which has the Na+ binding signature on its csubunits (Rahlfs and Müller, 1997). These csubunits are also assembled into a very stable complex, which probably indicates that intersubunit bridging by Na+ is the reason for the high stability of this assembly as well. Further in accordance with the role of Na+ in stabilizing the oligomeric ring structure are the properties of cE65 mutants. After exchanging this important Na+ binding residue by other amino acids, stable undecameric c-rings could no longer be isolated.
Figure 3.
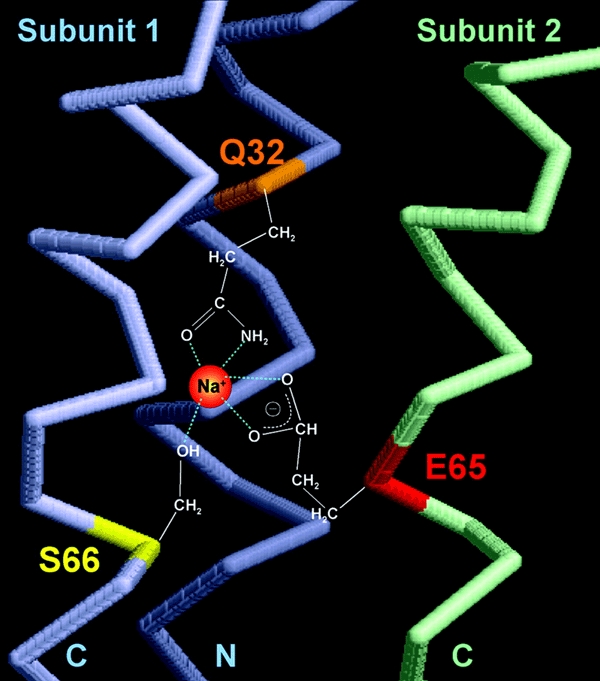
Structure model showing details of the Na+ coordination site in the I. tartaricus c11 ring. One subunit with C- and N-terminal α-helices is shown in light blue (subunit 1) and the adjacent C-terminal α-helix from the adjacent subunit in light green (subunit 2). The Na+ binding pocket is coordinated by at least three amino acid side chains. Subunit 1 provides glutamine 32 (green) and serine 66 (yellow) located on the N- (N) and C-terminal (C) helix, respectively. Glutamate 65 (red) is located on the C-terminal helix of subunit 2.
The results of this study imply that the 11 binding sites on c11 are accessible for Na+ or Li+ without a contribution from another subunit, in accordance with the 1a+11c channel model (von Ballmoos et al., 2002). They further show that occupancy of these sites is in dissociation equilibrium with the Na+ or Li+ concentration in the environment. The same applies for csubunit sites outside the a-subunit interface in the native ATP synthase complex. At the a-subunit interface, however, the c-subunit intrinsic channels must be closed, and Na+, Li+ or H+ can bind to rotor sites or dissociate from them only via the a-subunit channel to the opposite surface of the membrane.
The hypothesis that salt bridges between positively and negatively charged amino acids of adjacent monomeric units contribute to the ring stability was checked by mutagenesis of one charged residue of a putative pair against a neutral one. Three salt pairs, D2–K7, K50–D52 and K38–E41, have been proposed to stabilize the I. tartaricus c11 ring (Vonck et al., 2002). These salt pairs are only rarely found in csubunits with reduced temperature stability. The mutant D52L has little effect on the ring stability, which indicates that a K50–D52 pair is not a determinant for the stability of the c-oligomer from P. modestum. In contrast, no stable c-ring could be isolated from the D2C mutant. This result would be compatible with a role of the D2–K7 pair in the ring stability, but further experiments have to be performed to validate this hypothesis. The contribution of salt bridges to the ring stability is in accordance with its dependence on pH. Whereas stable ring structures persisted over the pH range from 4.5 to 11.5, the assembly was increasingly destabilized at higher or lower pH values. As acidic amino acids become protonated at the low pH values, and as basic amino acids became deprotonated at the high pH values, the salt bridges are obviously broken under these conditions. Furthermore, protonation of cE65 at acidic pH abolishes the Na+ binding site, which causes further destabilization of the c-ring. However, predictions of the heat stability of a c-ring on the basis of primary sequence comparisons from thermostable and thermolabile c-subunit oligomers are misleading and not conclusive enough at this stage.
The rationale for the stability of proteins has been intensively studied in the past (Robertson and Murphy, 1997). It is now generally believed that there is a set of molecular features that contribute to the heat tolerance of a protein rather than a specific molecular design (Vieille and Zeikus, 1996; Jaenicke and Bohm, 1998). This set includes hydrogen bonding, ionic stabilization of α-helices, tighter packing of the protein core, increased number of salt links and salt-link networks, increased number of disulfide bonds, decreased conformational entropy of the unfolded state and increased aromatic stacking interactions. In this paper, we show for the first time that a protein complex obtains an unusual heat tolerance due to an ion crossbridging mechanism.
Methods
Materials.
Chemicals were purchased from Fluka (Buchs, Switzerland). N-lauroylsarcosine sodium salt and n-octyl-β-D-glucopyranoside were purchased from Sigma and Glycon Biochemicals, respectively. Primers were custom synthesized by Microsynth (Balgach, Switzerland). Endogenous Na+ was minimized using chemicals with low Na+ content throughout all preparation procedures.
Purfication of I. tartaricus c11 ring from ATP synthase was performed as described elsewhere (T. Meier et al., in preparation).
Plasmid pt7c (Matthey et al., 1997) was mutagenized (Quik Change Site Directed Mutagenesis Kit, Stratagene) to yield the single mutations D2C, D52L, E65C, E65D and E65Q in the P. modestum csubunit.
Expression and purification of P. modestum c-oligomer from the strains BL21.
Strain E. coli BL21(DE3) (Novagen) was transformed with different plasmids described above. The transformed E. coli cells were grown in 2 l of LB medium to OD 0.6 at 37°C; after cooling on ice for 5 min, they were induced with 0.7 mM isopropylthiogalactoside. Expression was performed for 3–21 h at 30°C. Cells were harvested and frozen for storage at –70°C. Thawing of the cells was performed on ice after suspension in 5 ml of 10 mM potassium phosphate buffer pH 8.0 containing 5 mM ethylene diamine tetra-acetate (EDTA), 1 mM 1,4-dithio-DL-threitol and 0.1 mM diisopropylfluoro-phosphate per gram (wet weight) of cells. Preparation of membranes was performed at 4°C. Usually 0.5–2 g of cells were passed twice through a French pressure cell at 12 000 p.s.i. (8.3 × 104 kPa). After removal of cell debris by centrifugation at 15 000 g for 15 min, ultracentrifugation was performed at 200 000 g for 60 min. The membrane pellet was washed once with 5 ml of 10 mM Tris-HCl pH 8.0 containing 5 mM EDTA. The membranes were solubilized with 5 ml per gram cells of 20 mM Tris–HCl pH 8.0 containing 5 mM EDTA and 1% N-lauroylsarcosine for 10 min at 65°C. After ultracentrifugation at room temperature, the pellet was discarded and contaminating membrane proteins were precipitated with 65% (NH4)2SO4. After 20 min incubation at 25°C, the sample was centrifuged for 15 min at 39 000 g. The filtered supernatant containing the c-oligomer was dialyzed against 10 mM Tris–HCl pH 8.0. Stability of the purified c-oligomer was checked by SDS–PAGE (Schägger and Jagow, 1987). The gels were stained with silver (Nesterenko et al., 1994).
Stability assay for the c-oligomer from the ATP synthase.
The stability of the c-oligomer of the ATP synthase from I. tartaricus strain DSM 2382 (Schink, 1984), and the recombinant P. modestum protein, expressed in BL21(DE3), was tested by incubation of 1 μg of c-oligomer in different buffers: 50 mM citric acid–KOH pH 3.0–6.0; 50 mM Tris–HCl pH 6.0–9.0; 50 mM boric acid–KOH pH 9.0–11.0; and 5 mM KCl–KOH pH 11.0–13.0 and 1% octylglucoside or 0.1% N-lauroylsarcosine. These buffers also contained NaCl or LiCl in the range 0–100 mM. The incubation temperature was 95°C, and the incubation time varied from 0 to 60 min.
Acknowledgments
The authors thank Fabienne Henzen for excellent technical assistance. Franziska Wehrle is acknowledged for providing the mutant plasmids pt7c(E65D) and pt7c(E65Q).
References
- Abrahams J.P., Leslie A.G.W., Lutter R. and Walker J.E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature, 370, 621–628. [DOI] [PubMed] [Google Scholar]
- Aufurth S., Schägger H. and Müller V. (2000) Identification of subunits a, b, and c1 from Acetobacterium woodii Na+-F1F0-ATPase. Subunits c1, c2, and c3 constitute a mixed c-oligomer. J. Biol. Chem., 275, 33297–33301. [DOI] [PubMed] [Google Scholar]
- Boyer P.D. (1993) The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta, 1140, 215–250. [DOI] [PubMed] [Google Scholar]
- Dimroth P., Wang H., Grabe M. and Oster G. (1999) Energy transduction in the sodium F-ATPase of Propionigenium modestum. Proc. Natl Acad. Sci. USA, 96, 4924–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke R. and Bohm G. (1998) The stability of proteins in extreme environments. Curr. Opin. Struct. Biol., 8, 738–748. [DOI] [PubMed] [Google Scholar]
- Kaim G. and Dimroth P. (1999) ATP synthesis by F-type ATP synthases is obligatorily dependent on the transmembrane voltage. EMBO J., 18, 4118–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaim G., Wehrle F., Gerike U. and Dimroth P. (1997) Molecular basis for the coupling ion selectivity of F1F0 ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry, 36, 9185–9194. [DOI] [PubMed] [Google Scholar]
- Kaim G., Matthey U. and Dimroth P. (1998) Mode of interaction of the single a subunit with the multimeric c subunits during the translocation of the coupling ions by F1F0 ATPases. EMBO J., 17, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaim G., Prummer M., Sick B., Zumofen G., Renn A., Wild U.P. and Dimroth P. (2002) Coupled rotation within single F0F1 enzyme complexes during ATP synthesis or hydrolysis. FEBS Lett., 525, 125–163. [DOI] [PubMed] [Google Scholar]
- Kluge C. and Dimroth P. (1993a) Specific protection by Na+ or Li+ of the F1F0-ATPase of Propionigenium modestum from the reaction with dicyclohexylcarbodiimide. J. Biol. Chem., 268, 14557–14560. [PubMed] [Google Scholar]
- Kluge C. and Dimroth P. (1993b) Kinetics of inactivation of the F1F0 ATPase of Propionigenium modestum by dicyclohexylcarbodiimide in relationship to H+ and Na+ concentration: probing the binding site for the coupling ions. Biochemistry, 32, 10378–10386. [DOI] [PubMed] [Google Scholar]
- Laubinger W. and Dimroth P. (1987) Characterization of the Na+stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0 type. Eur. J. Biochem., 168, 475–480. [DOI] [PubMed] [Google Scholar]
- Matthey U., Kaim G. and Dimroth P. (1997) Subunit c from the sodium-ion-translocating F1F0-ATPase of Propionigenium modestum. Production, purification and properties of the protein in dodecylsulfate solution. Eur. J. Biochem., 247, 820–825. [DOI] [PubMed] [Google Scholar]
- Nesterenko M.V., Tilley M. and Upton S.J. (1994) A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods, 28, 239–242. [DOI] [PubMed] [Google Scholar]
- Neumann S., Matthey U., Kaim G. and Dimroth P. (1998) Purification and properties of the F1F0 ATPase of Ilyobacter tartaricus, a sodium ion pump. J. Bacteriol., 180, 3312–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji H., Yasuda R., Yoshida M. and Kinosita K. (1997) Direct observation of the rotation of F1-ATPase. Nature, 386, 299–302. [DOI] [PubMed] [Google Scholar]
- Rahlfs S. and Müller V. (1997) Sequence of subunit c of the Na+-translocating F1F0 ATPase of Acetobacterium woodii: proposal for determinants of Na+ specificity as revealed by sequence comparisons. FEBS Lett., 404, 269–271. [DOI] [PubMed] [Google Scholar]
- Robertson A.D. and Murphy K.P. (1997) Protein structure and the energetics of protein stability. Chem. Rev., 97, 1251–1267. [DOI] [PubMed] [Google Scholar]
- Schägger H. and Jagow G. (1987) Tricinesodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schink B. (1984) Fermentation of tartrate enantiomers by anaerobic bacteria, and description of two new species of strict anaerobes, Ruminococcus pasteurii and Ilyobacter tartaricus. Arch. Microbiol., 139, 409–414. [Google Scholar]
- Seelert H., Poetsch A., Dencher N.A., Engel A., Stahlberg H. and Müller D.J. (2000) Proton-powered turbine of a plant motor. Nature, 405, 418–419. [DOI] [PubMed] [Google Scholar]
- Stahlberg H., Müller D.J., Suda K., Fotiadis D., Engel A., Meier T., Matthey U. and Dimroth P. (2001) Bacterial Na+-ATP synthase has an undecameric rotor. EMBO rep., 2, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock D., Leslie A.G.W. and Walker J.E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science, 286, 1700–1705. [DOI] [PubMed] [Google Scholar]
- Vieille C. and Zeikus J.G. (1996) Thermozymes—identifying molecular determinants of protein structural and functional stability. Trends Biotechnol., 14, 183–190. [Google Scholar]
- von Ballmoos C., Appoldt Y., Brunner J., Granier T., Vasella A. and Dimroth P. (2002) Membrane topography of the coupling ion binding site in Na+-translocating F1F0 ATP synthase. J. Biol. Chem., 277, 3504–3510. [DOI] [PubMed] [Google Scholar]
- Vonck J., Krug von Nidda T., Meier T., Matthey U., Mills D.J., Kühlbrandt W. and Dimroth P. (2002) Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol., 321, 307–316. [DOI] [PubMed] [Google Scholar]


