Abstract
For DNA differential diagnosis of human Taenia cestodes, a base excision sequence scanning thymine-base method using the cytochrome c oxidase subunit I and cytochrome b genes as targets was used. The characteristic thymine-base peak profiles provide four distinct types, unique for T. saginata, T. asiatica, and two genotypes of T. solium. This approach provides a useful tool for the identification and diagnosis of human taeniid cestodes without DNA sequencing if nucleotide sequence databases are available.
Taenia solium and Taenia saginata are well-known parasites of medical and economic importance, causing cysticercosis in pigs and cattle and taeniasis in humans. T. solium is also an organism severely pathogenic to humans, causing fatal neurocysticercosis when cysticerci, the larval stage of T. solium, develop in the central nervous system. In recent years, this disease has been reported to be of particular importance not only in many developing countries (6, 15), but also in areas of nonendemicity, affecting immigrants, tourists (4), and, probably, refugees. Both T. solium and T. saginata have a worldwide distribution, but another human taeniid tapeworm, Taenia asiatica, which is phylogenetically and morphologically related to T. saginata, is distributed in Asian regions (5). This tapeworm was originally described as T. saginata subsp. asiatica. Interestingly, the intermediate host for T. asiatica is swine, as in the case of T. solium. In Asian regions, where these human taeniid cestodes are distributed sympatrically, it is not always possible to identify or characterize these cestodes accurately, including the cysticerci and eggs. In order to overcome limitations in identification of taeniid cestodes based on traditional methods, molecular approaches such as PCR-restriction fragment length polymorphism, single-strand conformation polymorphism (SSCP), and PCR have been reported (1, 7-9). Each of these methods has its advantages. More excitingly, it has been reported that T. solium forms two phylogenetic clusters—Asian and American/African groups—based on mitochondrial gene analysis (12-14), indicating the existence of two genotypes. This makes the morphological identification of human Taenia tapeworms more difficu lt.
In the present study, therefore, a base excision sequence scanning thymine-base (BESS T-base) system has been introduced for the accurate characterization of human Taenia and the two genotypes of T. solium. This system originally was developed for the identification of genetic variations (10, 11) and then was applied to virus isolates (3) and tumor suppressor genes (2). It detects mutations by incorporating limiting amounts of dUTP into a PCR product, resulting in the removal of the uracil base and cleavage of the phosphodiester bond at these abasic sites to produce a DNA ladder virtually identical to a thymine (T) sequencing ladder.
A total of 38 human taeniid samples were examined (Table 1). All parasite samples were stored in absolute ethanol at −30°C after collection. Mitochondrial DNA was prepared from an individual cysticercus or proglottid by using a DNeasy Tissue kit (Qiagen, Hilden, Germany). As target genes, the cytochrome c oxidase subunit I (Cox 1) and cytochrome b (Cytb) genes were used, because their nucleotide sequences have been determined completely (13). On the basis of the nucleotide sequences of the Cox 1 and Cytb genes from human Taenia cestodes, species- or genotype-specific T bases are dispersed over the genes. In this study, some regions containing characteristic T bases useful for differentiating among human Taenia cestodes were selected. First, the entire Cox 1 and Cytb genes were amplified for use as template DNA in BESS T-base analysis with Cox 1/F and Cox 1/R primers and Cytb/F and Cytb/R primers, respectively (13). For BESS T-base analysis, the following primer sets were utilized: P1(5′-ATATTTACTTTAGATCATAAGCG-3′, corresponding to nucleotide positions 28 to 50) and P2(5′-TAAAATTAATAGAACTAAAAAT-3′, positions 496 to 474) for Cox 1 (AB066485, AB066487-AB066491, and AB066493-AB066495 in Table 1) and P3(5′-TTATGAGATTGTCAAAAGATTCTT-3′, positions 170 to 193) and P4(5′-TATAGATGTCAAAACAGTAGCAGCCC-3′, positions 420 to 395) for Cytb (AB066570, AB066572-AB066576, AB066578, and AB066580-AB066581 in Table 1). The 5′ ends of the forward primers (P1 and P3) were labeled with the fluorescent dye 6-FAM. PCR for BESS T-base analysis was performed with the Epicentre Technologies BESS-T Base Reader kit. It had been confirmed that any of the buffers A, D, G, or J provided with the BESS-PCR opitimization kit (Epicentre Technologies) was suitable for BESS-PCR, because nonspecific PCR products were not amplified. AmpliTaq DNA polymerase (Applied Biosystems, Perkin-Elmer) without proofreading activity was used. The PCR protocols consisted of 30 cycles of denaturation (30 s at 94°C), annealing (30 s at 55°C), and extension (1 min at 72°C plus one cycle of 5 min at 72°C) for Cox 1 fragments. For the Cytb fragments, annealing was performed for 45 s at 52°C. PCR products (469 bp for the Cox 1 gene and 253 bp for the Cytb gene [data not shown]) were excised with uracil N-glycosylase and then cleaved with endonuclease IV. Subsequently, the samples were electrophoresed for 2.5 h in a 6% polyacrylamide sequencing gel containing 8 M urea. GeneScan 400HD (Applied Biosystems, Inc.) was used as a size standard.
TABLE 1.
Taeniid cestode samples used in this study
| Species | Developmental stage (no. tested) | Locality | Database accession no.
|
|
|---|---|---|---|---|
| Cox 1 gene | Cytb gene | |||
| T. saginata | Cysticercus (1) | Brazil | ||
| Cysticercus (1) | China (Urmuqi) | AB066495 | AB066581 | |
| Proglottid (1) | Indonesia | |||
| Proglottid (1) | Ethiopia | |||
| Proglottid (1) | Ecuador | |||
| Cysticercus (1) | Belgium | |||
| T. asiatica | Cysticercus (1) | Taiwan | AB066494 | AB066580 |
| Proglottid (1) | Taiwan | |||
| Cysticercus (1) | Korea | |||
| T. solium | Cysticercus (2) | China (Jilin) | AB066485 | AB066570 |
| Proglottid (1) | China (Yunnan) | AB066486 | AB066571 | |
| Proglottid (9) | China (Sichuan) | AB066486 | AB066571 | |
| Cysticercus (1) | Korea | |||
| Cysticercus (1) | India | AB066489 | AB066574 | |
| Cysticercus (1) | India (Vellore) | AB066489 | AB066574 | |
| Cysticercus (1) | India (Panjab) | AB066489 | AB066574 | |
| Cysticercus (1) | Thailand | AB066487 | AB066572 | |
| Cysticercus (1) | Indonesia | AB066488 | AB066573 | |
| Cysticercus (2) | Mozambique | AB066493 | AB066578 | |
| Cysticercus (1) | Tanzania | AB066493 | AB066578 | |
| Cysticercus (1) | Cameroon | AB066490 | AB066575 | |
| Cysticercus (2) | Ecuador | AB066491 | AB066576 | |
| Cysticercus (1) | Bolivia | AB066491 | AB066576 | |
| Cysticercus (1) | Brazil | AB066492 | AB066577 | |
| Cysticercus (2) | Mexico | AB066490 | AB066575 | |
| Cysticercus (1) | Peru | AB066490 | AB066575 | |
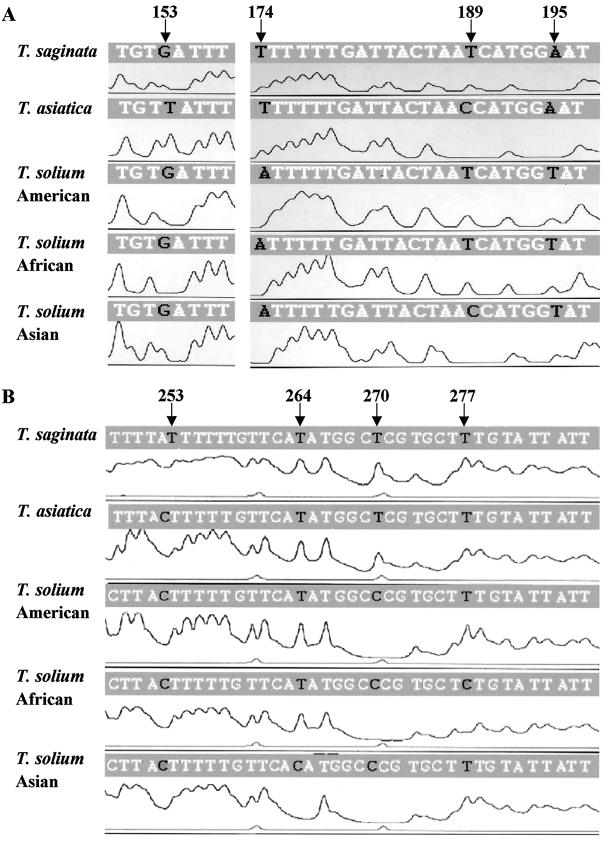
Based on the nucleotide sequences of Cox 1 from Taenia tapeworms, the bases at four positions (153, 174, 189, and 195) within the first 200 bp of the Cox 1 P1/P2 fragment are not conserved among human taeniid cestodes and thus seem to be useful as diagnostic markers (Fig. 1A). Figure 1 shows representative results of BESS T-base analysis. As expected, the T-base peak profiles could be categorized into four types unique for T. saginata, T. asiatica, and the two genotypes of T. solium. For example, nucleotide 153 of the Cox 1 gene is T only in T. asiatica, and is guanine in T. saginata and T. solium. The results of BESS T-base analysis were as expected. Namely, a T-base peak appeared at position 153 in T. asiatica, but no T-base peak appeared in the other Taenia tapeworms (Fig. 1A), demonstrating that the T-base peak serves as a differential marker. If the parasite is T. saginata, diagnostic T-base peaks will appear at both positions 174 and 189. Similarly, if T-base peaks appear at both positions 189 and 195, the taeniid parasites should be of the American/African genotype of T. solium. However, if a T-base peak appears at position 195, but none is present at position 189, the parasite can be identified as of the Asian genotype of T. solium. When the Cox 1 gene is used, the American/African genotype can be distinguished from the Asian genotype of T. solium, but it is impossible to differentiate between the American and African types of T. solium. By utilizing a Cytb fragment (P3/P4), however, it is possible to differentiate between the American and African types. In this Cytb fragment, four positions used as diagnostic markers are shown in Fig. 1B. T-base peaks appear at both positions 264 and 277 in the American type of T. solium, whereas a T-base peak at position 264, but not at position 277, indicates African T. solium. The Asian genotype of T. solium is easily identified by the lack of a T-base peak at position 264, because Asian T. solium has a cytosine at this position. The T-base peak profiles of T. saginata and T. asiatica are almost the same, but an additional T-base peak at position 253 appears in T. saginata because of the presence of six continuous T bases. Thus, by comparing characteristic T-base peak profiles, it is possible to differentiate easily and accurately among human taeniid cestodes without the need for DNA sequencing. SSCP is also a useful approach that can detect single-nucleotide differences without DNA sequencing, and it has been applied to the identification of human taeniid tapeworms (8), but the electrophoresis and autoradiography procedures involved are time-consuming. Compared with SSCP analysis, BESS T-base analysis is a much more rapid and facile protocol. In the BESS T-base system, mitochondrial DNA can be used directly as a template DNA instead of the entire Cox 1 and Cytb genes (data not shown). Furthermore, by utilizing a capillary system (for example, model ABI 310), it is also possible to shorten the analysis time.
FIG. 1.
BESS-T Base Reader analysis with Cox 1 (A) and Cytb (B) fragments. Profiles from T. saginata (China), T. asiatica (Taiwan), and T. solium (Brazil, Tanzania, and China) are shown. The nucleotide sequences indicated above the peaks are from the databases listed in Table 1. Diagnostic positions are denoted by arrows.
In conclusion, if nucleotide databases are available, the BESS T-base system provides a very useful tool, not only for the identifying taeniid specimens expelled from taeniasis patients, in order to speculate on the basis of molecular epidemiology and the locality where the patient was infected, but also for studying genetic variations in a variety of pathogenic agents and genes related to the diseases.
Acknowledgments
We thank T. Ikejima, D. C. Qiu, P. Dekumyoy, S. S. Margono, S. P. Sinha Babu, A. Oommen, G. Singh, P. C. Fan, K. Eom, V. C. W. Tsang, A. Plancarte, P. S. Craig, W. Benitez, C. M. Nunes, M. Vilhena, S. Geerts, A. A. Kassuku, S. S. Afonso, A. Zoli, and S. Miura for kindly providing parasite materials. This work was supported in part by Grants-in-Aid for Scientific Research to A.I. (no. 10557029, 11694259, and 12557024), M.N. (no. 12670228), and Y.S. (no. 12770122) from the Ministry of Education, Science, Sports, and Culture of Japan and the Japan Society for Promotion of Science.
REFERENCES
- 1.Bowles, J., and D. P. McManus. 1994. Genetic characterization of the Asian Taenia, a newly described taeniid cestode of humans. Am. J. Trop. Med. Hyg. 50:33-44. [PubMed] [Google Scholar]
- 2.Brieger, J., E. J. Weidt, K. Gansen, and H. J. Decker. 1999. Detection of a novel germline mutation in the von Hippel-Lindau tumor-suppressor gene by fluorescence-labelled base excision sequence scanning (F-BESS). Clin. Chem. 45:1564-1567. [DOI] [PubMed] [Google Scholar]
- 3.Charrel, R. N., N. Lévy, R. B. Tesh, and L. J. Chandler. 1999. Use of base excision sequence scanning for detection of genetic variations in St. Louis encephalitis virus isolates. J. Clin. Microbiol. 37:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Earnest, M. P., L. B. Reller, C. M. Filley, and A. J. Grek. 1987. Neurocysticercosis in the United States: 35 cases and a review. Rev. Infect. Dis. 9:961-979. [DOI] [PubMed] [Google Scholar]
- 5.Fan, P. C. 1989. Taiwan Taenia and taeniasis. Parasitol. Today 4:86-88. [DOI] [PubMed] [Google Scholar]
- 6.Flisser, A. 1988. Neurocysticercosis in Mexico. Parasitol. Today 4:131-137. [DOI] [PubMed] [Google Scholar]
- 7.Gasser, R. B., and N. B. Chilton. 1995. Characterisation of taeniid cestode species by PCR-RFLP of ITS2 ribosomal DNA. Acta Tropica 59:31-40. [DOI] [PubMed] [Google Scholar]
- 8.Gasser, R. B., X. Zhu, and W. Woods. 1999. Genotyping Taenia tapeworms by single-strand conformation polymorphism of mitochondrial DNA. Electrophoresis 20:2834-2837. [DOI] [PubMed] [Google Scholar]
- 9.González, L. M., E. Montero, L. J. S. Harrison, R. M. E. Parkhouse, and T. Garate. 2000. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J. Clin. Microbiol. 38:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkins, G. A., and L. M. Hoffman. 1997. Base excision sequence scanning. Nat. Biotechnol. 15:803-804. [DOI] [PubMed] [Google Scholar]
- 11.Hawkins, G. A., and L. M. Hoffman. 1999. Rapid DNA mutation identification and fingerprinting using base excision sequence scanning analysis. Electrophoresis 20:1171-1176. [DOI] [PubMed] [Google Scholar]
- 12.Ito, A., M. Nakao, M. Okamoto, Y. Sako, and H. Yamasaki. 2002. Mitochondrial DNA of Taenia solium: from basic to applied science, p. 47-55. In S. Prabhakar and G. Singh (ed.), Taenia solium cysticercosis. CAB International, Wallingford, United Kingdom.
- 13.Nakao, M., M. Okamoto, Y. Sako, H. Yamasaki, K. Nakaya, and A. Ito. 2002. A phylogenetic hypothesis for the distribution of two genotypes of the pig tapeworm Taenia solium worldwide. Parasitology 124:657-662. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto, M., M. Nakao, Y. Sako, and A. Ito. 2001. Molecular variation of Taenia solium in the world. Southeast Asian J. Trop. Med. Public Health 32(Suppl. 2):90-93. [PubMed] [Google Scholar]
- 15.Simanjuntak, G. M., S. S. Margono, M. Okamoto, and A. Ito. 1997. Taeniasis/cysticercosis in Indonesia as an emerging disease. Parasitol. Today 13:321-323. [Google Scholar]