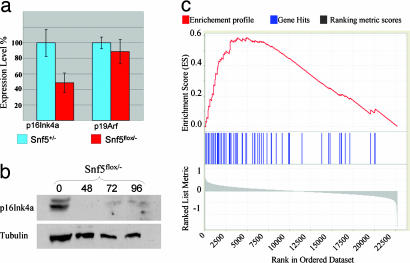
Fig. 4.
Loss of Snf5 leads to down-regulation of p16Ink4 and up-regulation of E2F target genes. (a) Quantitative real-time PCR analysis of p16Ink4a and p19Arf at 72 h after adenovirus-Cre transduction. (b) Anti-p16Ink4a Western blot of protein from Snf5flox/- cells at 0, 48, 72, and 96 h after adenovirus-Cre transduction. (c) E2F target genes are up-regulated after inactivation of Snf5. Each vertical blue line represents an E2F target as defined by the work of Vernell et al. (18). The left-to-right position of each line indicates the relative position of the probe within the rank ordering of the 22,690 probes present on the affy430A2.0 mouse microarray from the probe most up-regulated after Snf5 inactivation (position 1 on the left) to the most down-regulated (position 22690 on the right). The genes near the middle are unaffected by Snf5 loss. The E2F target gene set is clearly enriched among genes up-regulated in the MEF Snf5 data set, as evidenced by the increased number of blue marks on the left side of the distribution and the positive enrichment score marked by the red line (P < 1 × 10-4).