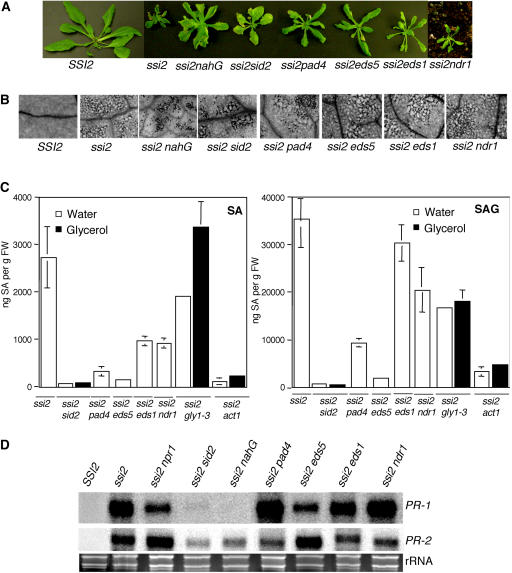
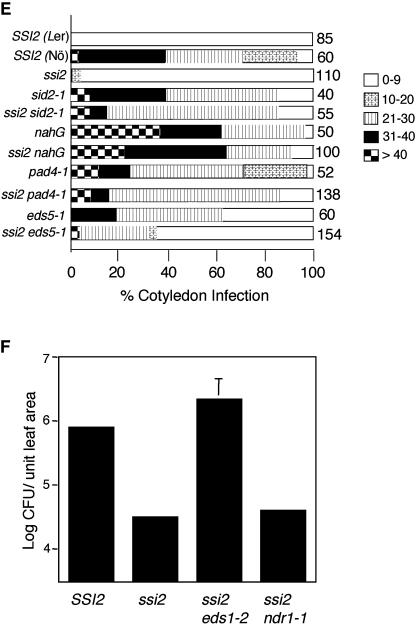
Figure 2.
Morphological, molecular, and biochemical phenotypes of wild-type, ssi2, ssi2 nahG, ssi2 sid2, ssi2 pad4, ssi2 eds1, ssi2 eds5, and ssi2 ndr1 plants. A, Comparison of the morphological phenotypes displayed by the wild-type (SSI2, Nö ecotype), ssi2, and various double-mutant plants in the ssi2 background. B, Microscopy of trypan blue-stained leaves from wild-type (SSI2, Nö ecotype), ssi2, and various double-mutant plants in the ssi2 background. C, Endogenous SA and SAG levels in the leaves of indicated 4-week-old soil-grown plants treated with water or glycerol. The values are presented as the mean of three replicates. Error bars represent sd. D, Expression of the PR-1 and PR-2 genes in indicated genotypes. RNA gel-blot analysis was performed on 7 μg of total RNA extracted from 4-week-old soil-grown plants. SSI2 indicates Nö ecotype. Ethidium bromide staining of rRNA was used as a loading control. E, Growth of P. parasitica biotype Emco5 on various plant genotypes listed at the left. The Ler and Nö ecotypes were used as the resistant and susceptible controls, respectively. The numbers against each box indicate cotyledons scored. The shade of each box indicates the severity of infection, based on the number of sporangiophores per cotyledon (see key at the right). F, Growth of P. syringae on SSI2, ssi2, ssi2 eds1, and ssi2 ndr1. Four leaf discs were harvested from infected leaves at 3 d postinoculation, ground in 10 mm MgCl2, and the bacterial numbers tittered. The bacterial numbers ± sd (n = 4) presented as colony forming units (CFU) per unit leaf area of 25 mm2. The experiment was independently performed twice with similar results.