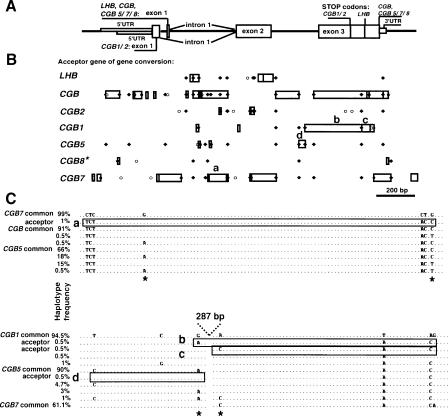
Figure 2.
(A) Consensus exon–intron structure of LHB/CGB genes presented in parallel with (B) the summary distribution of gene-conversion acceptor sites and multisite variants (MSVs) within LHB/CGB genes. A minimum gene-conversion site (□) was defined as a segment within an acceptor gene including ≥2 associated SNPs, for which a potential donor gene could be identified (detailed information in Table 3). Typically, for each gene-conversion site, several converted tracts could be determined (overlapping □). MSV1 (○) is defined as an SNP in one of the duplicate genes, which is also represented as a paralogous sequence variant among homologous genes (Fredman et al. 2004). MSV2 (♦) is an SNP that is present as a polymorphism at the identical sequence position in several duplicated genes. Several gene-conversion sites overlap with MSVs, indicating their origin from a gene-conversion event, rather than parallel mutation. Lettering (a, b, c, d) refers to gene-conversion sites, which are described in detail in C. CGB8* data are from the analysis of 11 Estonian individuals; the analysis of the rest of the genes is based on a combined sample set of 95 Estonian (n = 47), Han (n = 25), and Mandenka (n = 23) individuals. (C) Examples of gene-conversion segments (boxed sequences a–d) and their potential donor genes. Haplotype frequencies of each segment variant are derived from the combined sample (n = 95) of Estonians, Mandenka, and Han Chinese. Segment a within CGB7 (frequency 1%) originates most probably from a common variant of CGB (91%) or CGB5 (66%). Segments b (0.5%) and c (0.5%) within CGB1 were derived by gene conversion from common variants of CGB5 (90%) and CGB7 (61.1%), respectively. Reciprocally, a common variant of CGB1 (94.5%) or CGB7 (61.1%) is the most apparent donor of segment d to CGB5 (0.5%). “Shared SNPs” or MSV2 between gene pairs (*) are colocalized with gene-conversion sites.