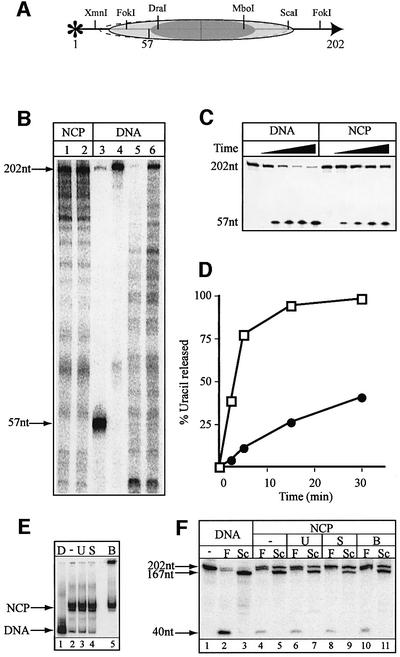
Fig. 5. Removal of uracil from nucleosome core particles reconstituted on a 202 bp DNA fragment. An extended fragment of the L.variegatus 5S rRNA gene (202 bp) containing a uracil residue at position 57 (numbered from the 5′ end of the 202 bp fragment) was constructed. (A) Cartoon showing the major and minor (dashed line) translational positions of the histone octamers based on digestion with the indicated restriction enzymes. (B) DNase I footprinting of core particles (lanes 1 and 2) shown next to the cleaved DNA substrate following UNG2 and piperidine treatment (lane 3) to verify that the uracil residue is positioned within DNA protected by octamers. The 202 bp DNA substrate (lane 4) and DNase I footprinting of the naked DNA (lanes 5 and 6) are shown for comparison. (C) An 8% polyacrylamide/7 M urea/20% formamide gel showing the time course of uracil excision by the UNG2 glycosylase from naked DNA (DNA) and nucleosome core particles (NCP). (D) Rates of uracil excision in naked DNA (open squares) and NCP (closed circles) after correction for uracil removal from naked DNA present in the core particle preparation (average of three independent experiments). The SEM of uracil removal was <20%. (E) Native 5% polyacrylamide gel after a 30 min incubation, showing naked DNA (D), and core particles incubated alone (–) or with UNG2 (U), SMUG1/APE1 (S) or BER proteins (B). (F) An 8% polyacrylamide/7 M urea/20% formamide gel showing restriction enzyme digestion of 202 bp naked DNA (lanes 1–3) with FokI (F) or ScaI (Sc) and, similarly, core particles alone (–) or after uracil removal by UNG2 (U), SMUG1/APE1 (S) or BER enzymes (B) after a 30 min incubation.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.