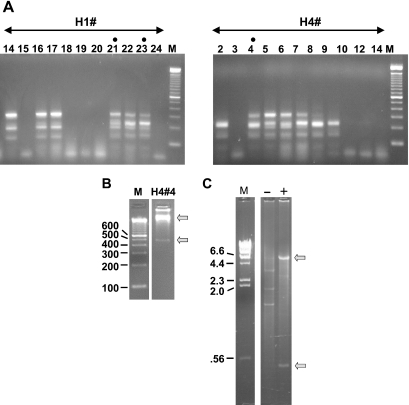
Figure 3.
(A) Colony PCR analysis of yeast clones obtained from H1 and H4 genomic DNA by the method shown in Figure 1B. Ligated samples (see legend, Figure 2) were used to transform S.cerevisiae. In contrast to results with E.coli (Figure 2), full-length candidate clones were obtained. Lanes with dots are from colonies used in further analyses (see text). The DNA ladder is as in Figures 1 and 2. (B) Verification of clone structure. To confirm the structure of the full-length candidates, yeast minipreps were treated with EcoR1 and subjected to random-primed rolling circle amplification with phi-29 DNA polymerase. As an example, H4#4 is displayed on a 2.5% agarose gel after diagnostic digestion with Xbal and PvuII. Upper and lower arrows indicate the vector backbone and insert bands, respectively. (C) Efficacy of the EcoR1 pre-digestion. The EcoR1 pre-treated sample in B (‘+’) is run on a 1% agarose gel alongside an untreated (‘−’) miniprep of H4#4. Treated and untreated samples were phi-29 amplified in parallel and digested with XbaI and PvuII. Bands evident in the untreated lane correspond in size to those predicted for 2 µ circle DNA. Bands observed with EcoR1 pre-treatment (arrows) correspond to the pYes2.1 vector and insert.