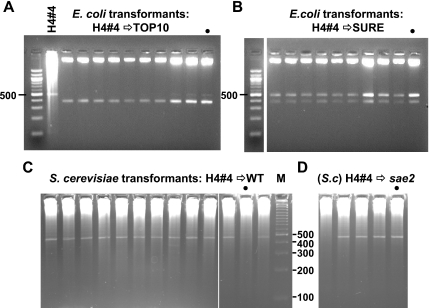
Figure 4.
Side-by-side comparison of microbial hosts for their ability to maintain the same plasmid. (A) Universal deletion in E.coli Top 10 cells. Plasmids re-isolated from TOP 10 clones transformed with the H4#4 plasmid are deleted. Each was XbaI and PvuII digested. H4#4 DNA (also cut with XbaI and PvuII after a phi-29 amplification) is loaded adjacent to the marker lane. Note, a different 100 bp ladder was used here (New England Biolabs) which has an intense 500 bp rather than 600 bp band as in previous figures. (B) Instability of the H4#4 plasmid in E.coli SURE cells. Plasmid DNA from individual SURE H4#4 transformants is a mixture of deleted and apparently non-deleted forms despite the lack of a functional SbcCD nuclease. The gel image was cut to remove one lane. (C) Stability of H4#4 plasmid in wild-type yeast. Phi-29 amplified minipreparations of DNA from random wild-type yeast clones transformed with H4#4 DNA were digested with XbaI and PvuII. Full-length inserts are observed. (D) Phi-29 amplified minipreparations of DNA from random sae2 yeast clones transformed and analyzed as in (C). In A–D, dots mark samples from colonies that were re-streaked as described in the text.