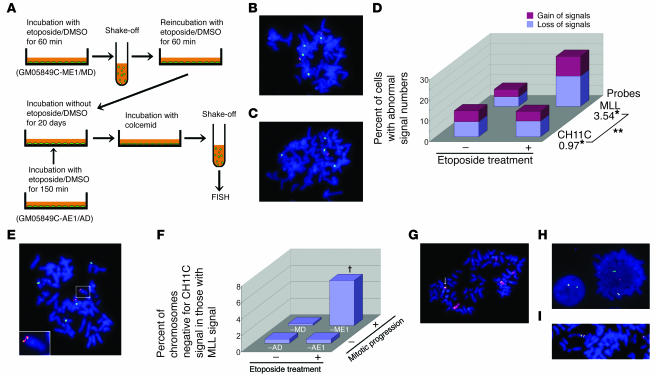
Figure 6.
Chromosomal aberrations in a mixture of stable clones of ATM-deficient fibroblasts, which had executed mitosis under etoposide treatment. (A) Flow diagram for long-term culture procedure. (B, C, and E) Metaphase spreads hybridized with the CH11C probe (green) and MLL probes (green and red overlap). (B) One GM05849C-MD cell showing 4 CH11C signals and 2 pairs of MLL signals. (C) One GM05849C-ME1 cell showing 4 CH11C signals with only 1 of them bearing MLL signals. (D) Proportion of cells with gain and loss of CH11C or MLL signals. n = 300 for each. *OR for etoposide treatment. **P = 0.0002, difference in OR for interaction term of probe × treatment by a logistic regression model. (E) One GM05849C-ME1 cell with the MLL gene translocated to another chromosome. The inset is a magnified image of the enclosed area. (F) Percent of chromosomes negative for CH11C signal among those positive for MLL signals. Data were analyzed by multiple logistic regression. n = 400 for each. †P = 0.028 for interaction term of etoposide treatment × mitotic progression. Neither of the pairwise comparisons for etoposide treatment in asynchronous cells nor for mitotic progression in etoposide-untreated cells was statistically significant (P = 1.0). (G) Representative image of chromosome 11 translocation (arrow) in GM05849C-ME1 cells hybridized with chromosome 11 painting (red) and MLL probes (green and red overlap). (H and I) Abnormal MLL gene configuration. GM05849C-ME1 cells were hybridized with MLL probes. Chromosomal translocation of MLL BCR (H) and tandem duplication of the MLL gene (I). Original magnification for FISH images, ×600.