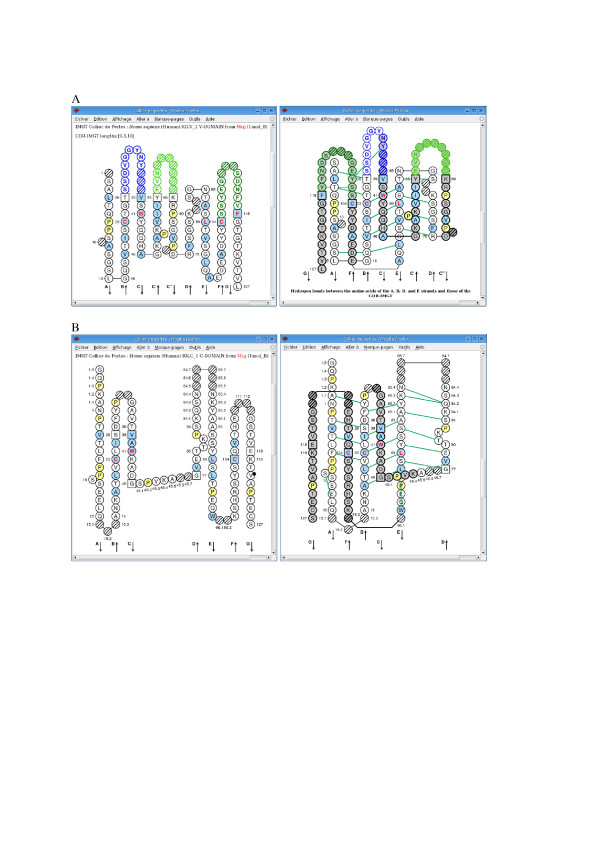
Figure 2.
IMGT Colliers de Perles of a V-DOMAIN (A) and of a C-DOMAIN (B) (code PDB 1mcd in IMGT/3Dstructure-DB [9]). IMGT Colliers de Perles are shown on one layer (on the left hand side) and on two layers with hydrogen bonds (on the right hand side). (A) The IMGT Collier de Perles of a V-DOMAIN is based on the IMGT unique numbering for V-DOMAIN and V-LIKE-DOMAIN [16]. The CDR-IMGT are limited by amino acids shown in squares, which belong to the neighbouring FR-IMGT. The CDR3-IMGT extends from position 105 to position 117. CDR-IMGT regions are colored as follows on the IMGT site: CDR1-IMGT (blue), CDR2-IMGT (bright green), CDR3-IMGT (dark green) and hydrogen bonds are shown as green lines. (B) The IMGT Collier de Perles of a C-DOMAIN is based on the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN [17]. Amino acids are shown in the one-letter abbreviation. Arrows indicate the direction of the beta strands that form the two beta sheets of the immunoglobulin fold [3, 4]. Hatched circles correspond to missing positions according to the IMGT unique numbering [16, 17]. In the IMGT Collier de Perles on the IMGT Web site http://imgt.cines.fr hydrophobic amino acids (hydropathy index with positive value) and Tryptophan (W) found at a given position in more than 50 % of analysed IG and TR sequences are shown in blue, and all Proline (P) are shown in yellow.