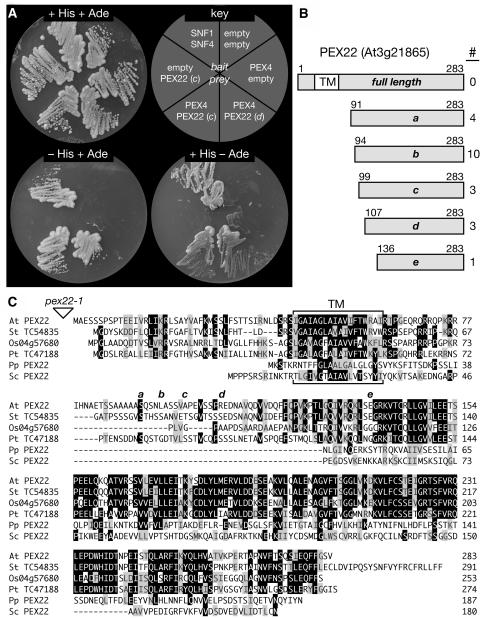
Figure 5.
PEX4 Interacts with Arabidopsis PEX22.
(A) A yeast two-hybrid assay with PEX4 fused to the GAL4 DNA binding domain (bait) was used to query an Arabidopsis cDNA library fused to the GAL4 activation domain (prey). The growth of the resulting yeast strains is shown on plates containing complete medium (top left) or under selective conditions indicating protein interaction (−His at bottom left, −adenine [Ade] at bottom right). Two PEX4-interactor strains are shown, SNF1 and SNF4 are positive controls (Fields and Song, 1989), and the empty vector combinations are negative controls.
(B) All 21 PEX4 interactors are PEX22 (At3g21865). The gray bar represents the PEX22 protein; a predicted transmembrane domain (TM) is indicated by the white box. The five identified clones are designated a to e, with the terminal amino acid numbers above the bar and the number of isolates indicated at right.
(C) Protein alignment of Arabidopsis (At) PEX22 with similar putative proteins from potato (Solanum tuberosum; St), rice (Os), and loblolly pine (Pinus taeda; Pt) and characterized yeast PEX22 proteins from S. cerevisiae (Sc) and P. pastoris (Pp). Potato and pine sequences are from genome projects assembled in The Institute for Genomic Research Gene Index Database (http://www.tigr.org/tdb/tgi; Quackenbush et al., 2001), and Os0457680 is from The Institute for Genomic Research Rice Genome Annotation (http://www.tigr.org/tdb/e2k1/osa1/index.shtml; Yuan et al., 2005). Sequences were aligned with the MEGALIGN program (DNASTAR) using the ClustalW method with BLOSUM series protein weight matrix. Residues identical or chemically similar in a majority of sequences are indicated by black or gray boxes, respectively. The T-DNA insertion disrupting pex22-1 upstream of the ATG is represented by the triangle. The predicted transmembrane domain (TM) is bracketed, and the starting amino acids of each of the five two-hybrid clones represented in (B) are indicated by a to e above the sequence.