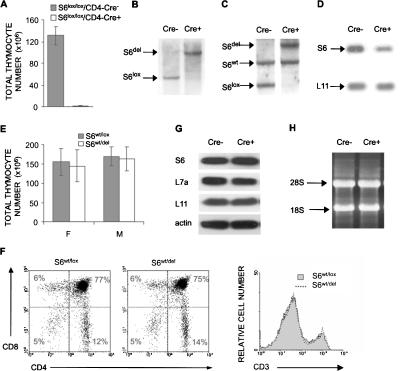
Figure 1.
Conditional deletion of the S6 gene in the thymus. (A) Total number of thymocytes in S6lox/lox/CD4-Cre- and S6lox/lox/CD4-Cre+ mice (n = 14 and 10, respectively). (B) Southern blot analysis of genomic DNA from pooled thymi of S6lox/lox/CD4-Cre+ and S6lox/lox/CD4-Cre- mice using S6 intron-specific probe. The positions of DNA fragments representing S6del and S6lox are shown. (C) Southern blot analysis of genomic DNA from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ thymi using S6 intron-specific probe. The positions of DNA fragments representing S6del, S6lox, and S6wt alleles are indicated. (D) Northern blot analysis of total RNA from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ thymi using S6 cDNA as a probe. The L11 cDNA probe was used as a control for loading the same quantity of total RNA on the gel. (E) Total number of thymocytes from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ mice. (F) S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ thymocytes were stained with CD4 and CD8 antibodies (left panel) or anti-CD3 antibodies (right panel), and analyzed by FACS. Cell surface markers are shown as coordinates. The numbers in the quadrant refer to the percentage of a particular T-cell population of live cells. (G) The protein expression of ribosomal proteins S6, L7a, and L11 in thymocytes from S6wt/lox/CD4-Cre+ and S6wt/lox/CD4-Cre- mice was analyzed by Western blot. Reprobing with antibodies to actin served as a loading control. (H) Total RNA from 15 × 106 S6wt/lox/CD4-Cre+ and S6wt/lox/CD4-Cre- thymocytes was separated in agarose gel containing formaldehyde, transferred to a nitrocellulose membrane, and stained with ethidium bromide. Positions of 28S and 18S rRNAs are shown.