Figure 2.
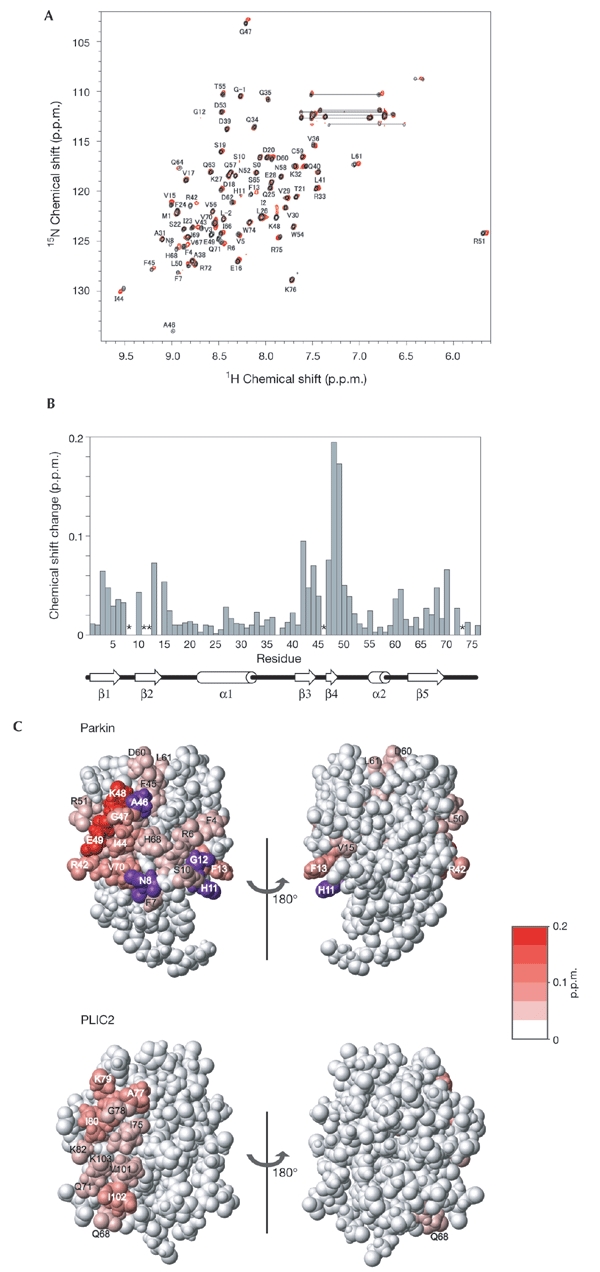
Identification of the binding site for Rpn10196–306 in the parkin ubiquitin-like (Ubl) domain. (A) 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of the parkin Ubl domain in the presence (red) and absence (black) of equimolar quantities of Rpn10196–306. The peaks labelled with L-2, G-1 and S0 originate from the amino-terminal tag. (B) NMR chemical-shift-perturbation data for the parkin Ubl domain. The data are displayed for each residue according to the equation (0.2 δN2 + δH2)1/2, where δN and δH represent the change in nitrogen and proton chemical shifts on addition of Rpn10196–306. Asterisks indicate residues the peaks of which became undetectable due to broadening. Secondary structure elements for the parkin Ubl are shown below the graph. (C) Mapping of the perturbed residues of the Ubl domains of parkin and PLIC2 (Walters et al., 2002) on binding to Rpn10. Residues showing a chemicalshift-perturbation are coloured in red, with the colour gradient indicating the strength of the perturbation. Residues the peaks of which became undetectable on binding to Rpn10 are shown in purple.
