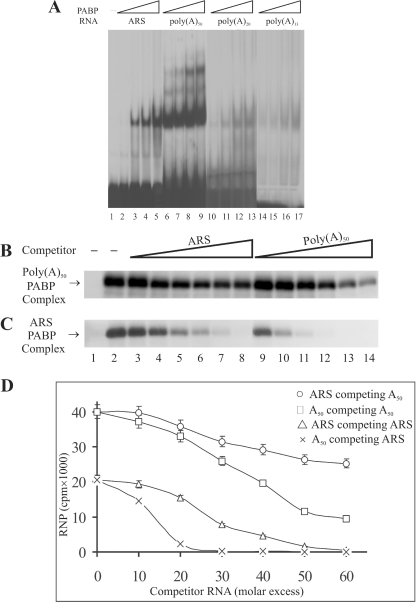
Figure 2.
Comparison of the ability of the ARS and poly(A) RNA to bind PABP. (A) Gel-shift assays of binding of PABP to the ARS and poly(A) RNAs of various length. REMSA was performed using 0.2, 0.4, 0.8 and 1.6 ng of purified 6× His-PABP and ≈1.5 ng of [32P]-labeled ARS (lanes 2–5), poly(A)50 (lanes 6–9), poly(A)20 (lanes 10–13) and poly(A)13 (lanes 14–17) RNAs. Samples were analyzed on 5% PAGE under non-denaturing conditions. (B and C) UV crosslinking assays of binding of PABP to ARS and poly(A)50 RNAs. The in vitro synthesized [32P]-labeled ARS and poly(A)50 RNAs (≈3 ng, 3 × 105) were incubated with the purified PABP (≈2 ng) for 5 min at room temperature. Unlabeled ARS (lanes 3–8) or poly(A)50 (lanes 9–14) competitor RNAs were added (10-fold molar excess increment) and incubated further at the room temperature for 3 min. Following the UV treatment, the samples were treated with RNase A/RNase T1, fractionated on a 10% SDS–PAGE and visualized by autoradiography. Lane 1, samples without UV treatment; lane 2, samples without unlabeled competitor RNA. (D) The RNP bands in (A and B) were excised by superimposing the radiograph and the level of radioactivity was measured by scintillation counter. The average level of radioactivity of the RNP complex in each band from three separate competition experiments was plotted against the molar concentration of the competitor RNA.