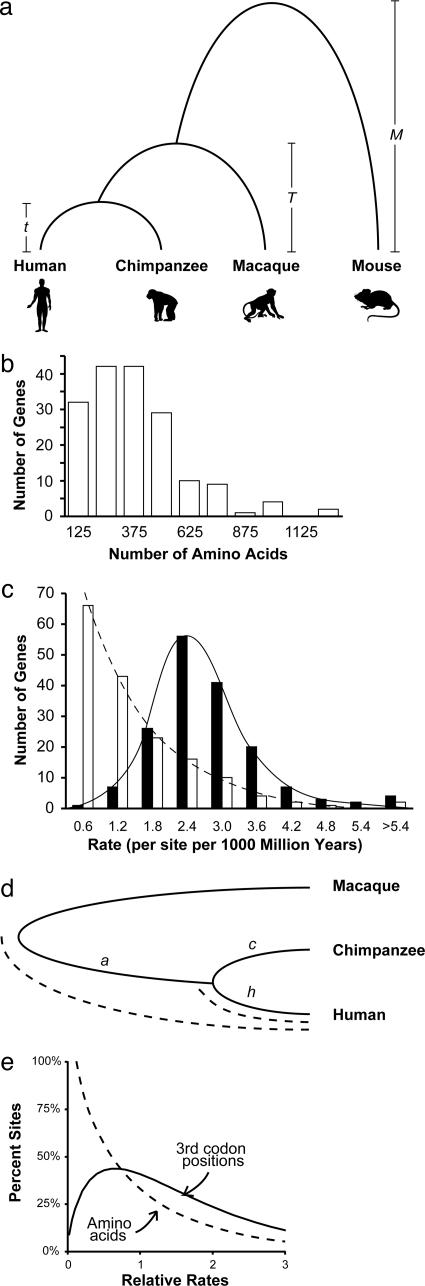
Fig. 1.
Characteristics of the data. (a) Phylogenetic relationships of human, chimpanzee, macaque, and mouse. (b) Histogram showing the length distribution of proteins used. (c) Distributions of the average evolutionary rates of amino acids (open bars) and third codon positions (solid bars) for 167 protein-coding genes analyzed in this study. Evolutionary rates were estimated assuming a 90 Ma date for primate–rodent divergence (5, 22, 46). (d) Schematic showing how lineage-specific molecular clocks were used to estimate the human–chimpanzee divergence time by using the ML distances between species pairs. The human–chimpanzee divergence time is given by the fraction ([h + c]/2)/(a + [h + c]/2) of the time assumed for ape–OWM divergence. (e) Distribution of evolutionary rates among sites for amino acids (dashed curve; shape parameter = 0.83) and third codon positions (solid curve; shape parameter = 1.65) obtained in ML analyses of concatenated sequences (53,008 codons) of 167 protein-coding genes. In each case, modeling the rate variation among sites by using a gamma distribution produced a significantly better fit (P < 0.005).