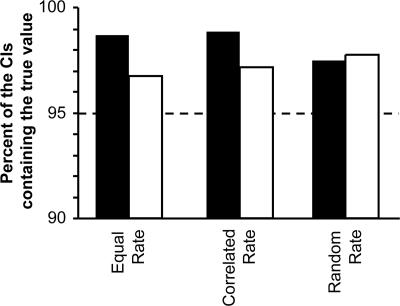
Fig. 2.
The percent of simulation replicates in which the 95% C.I. generated by the MBR method includes the simulated true value using the ML-distance (filled bars) and Bayesian approaches (open bars). (See Supporting Text for a description of the rate-variation models and how the data were parameterized.) We simulated 1,000 167-gene equivalent data sets under each rate-variation regime and conducted MBR analyses to obtain C.I.s for the ML-distance (GTR+Γ) and Bayesian (F84+Γ) approaches. The results show that 95% C.I.s contain the true value more often than required, particularly for the ML-distance method. This finding shows that modeling-rate variation among lineages (as in the Bayesian method) should lead to narrower C.I.s when the underlying model for correlated evolutionary rates among lineages is satisfied (constant and correlated rate cases), as compared with the ML-distance approach, in which rate variation is incorporated into the C.I. instead of being modeled. Interestingly but not unexpectedly, Bayesian and ML methods perform similarly when the assumptions of the Bayesian model are not satisfied (random rate-variation case). This finding is consistent with a study showing that a Bayesian approach with lognormal rate-variation model does not accommodate uncorrelated rate variation (47).