FIG. 2.
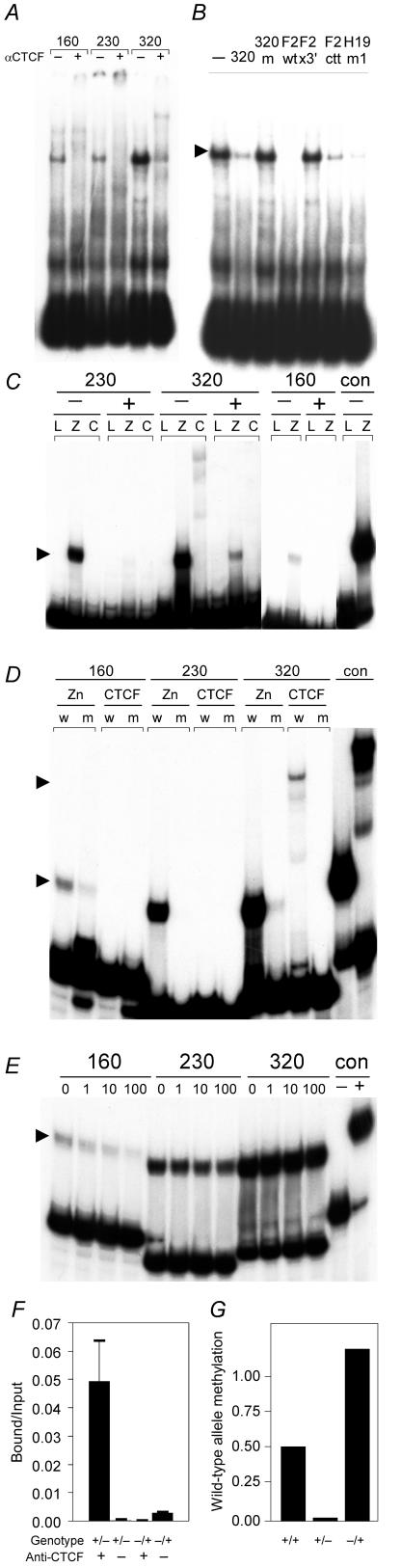
CTCF binds the unmethylated Rasgrf1 DMD. A. Oligonucleotide probes corresponding to three sequences in the DMD (230, 320, and 160) were tested for CTCF binding by gel shift analysis. Reactions included CTCF-containing extracts from chicken erythrocytes with (+) or without (−) anti-CTCF antibody. B. One of the probes (320) was used in further gel shift experiments using CTCF purified from chicken erythrocytes and several competitor probes. Binding reactions were done with no competing oligonucleotide (−) or 200-fold molar excesses of competitors that included self (320), methylated self (320m), the wild-type CTCF binding site at the chicken β-globin locus (F2wt), mutated forms of the chicken β-globin CTCF site that abolished (F2x3′) or attenuated (F2ctt) CTCF binding, and a CTCF site from H19. C. Methylation-sensitive binding was confirmed using larger probes containing the three Rasgrf1 CTCF sites prepared by PCR and methylated in vitro (+) or left unmethylated (−) prior to incubation with in vitro-transcribed and -translated luciferase (L), Zn-finger domain of CTCF (Z), or full-length CTCF (C). An unmethylated human XIST probe (con) was used as a positive control for binding. D. Probes generated by PCR corresponding to wild-type (w) or mutated (m) forms of Rasgrf1 CTCF sites 160, 230, and 320 were tested for binding to the in vitro-translated and -transcribed Zn-finger domain of CTCF. The mutated forms of these sites were those used in enhancer-blocking tests in Fig. 1. E. The wild-type probes from panel D were used in competition assays that included the Zn-finger domain of CTCF and increasing molar excesses (0 or 1-, 10-, or 100-fold) of unlabeled, mutated site probes. An unmethylated human XIST probe (con) was used with (+) or without (−) added protein as a control. Arrowheads in panels B, C, D, and E indicate positions of complexes with the full-length CTCF or Zn-finger domain complexes. F. Chromatin immunoprecipitation was performed using embryonic fibroblasts from mice with a paternally (+/−) or maternally (−/+) inherited repeat deletion (29) with (+) or without (−) antibody against CTCF. Precipitates were analyzed using real-time PCR with primers specific for the wild-type allele only. The experiment was done in triplicate, and the fraction of input that was precipitated is reported. Error bars show the standard deviation. G. The methylation state of HhaI sites in the wild-type allele was measured by real time PCR. DNAs from wild-type (+/+), +/−, and −/+ cells used in panel F were amplified with wild-type-specific primers that spanned five HhaI sites in the DMD. Amplification was done either before or after digestion with HhaI. Wild-type allele methylation is the ratio of product amplified from the digested templates to that from undigested templates. Of the two wild-type alleles in +/+ cells, only the paternal is methylated (ratio = 0.50), the single wild-type maternal allele in +/− cells is not appreciably methylated (ratio = 0.014), and the single wild-type paternal allele in −/+ cells is fully methylated (ratio = 1.2).