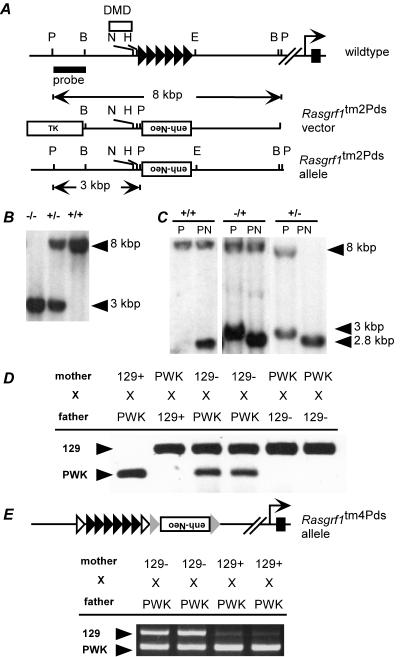
FIG. 3.
An extra enhancer at Rasgrf1 bypasses imprinted regulation caused by the DMD enhancer-blocking element. A. The wild-type Rasgrf1 locus was mutated to create a new allele (Rasgrf1tm2Pds) that placed an enhancer (enh-neo) in an inverted orientation at the locus in place of the repeats (filled triangles). B. Southern blot analysis using the probe shown in panel A and PstI-digested DNA revealed the expected 8.0-kbp and 3.0-kbp bands from the wild-type (+) and mutated (−) alleles, respectively, confirming homologous recombination. C. Methylation analysis by Southern blotting using PstI (P) and NotI (N) produced a 2.8-kbp band when methylation was absent from either the wild-type or mutated allele. Maternal transmission of the Rasgrf1tm2Pds allele (−/+) had no effect on methylation, while in mice with paternal transmission (+/−), the sole band at 2.8 kbp indicated that methylation of the paternal allele was lost. D. Allele-specific reverse transcriptase PCR using cDNA prepared from neonatal brains of progeny from reciprocal crosses between wild-type PWK mice and animals with the Rasgrf1tm2Pds mutation on the 129S4/SvJae (129) background. The extra enhancer-containing neo cassette facilitated expression of the otherwise silent maternal allele when it was maternally transmitted and enabled paternal allele expression even though paternal methylation was absent. E. Expression analysis of an independently derived allele with loxP sites (open triangles) flanking the repeats and frt sites (shaded triangles) flanking the enh-neo cassette (Rasgrf1tm4Pds) (R. Holmes et al., submitted) revealed the same escape from imprinted expression as for the Rasgrf1tm2Pds allele.