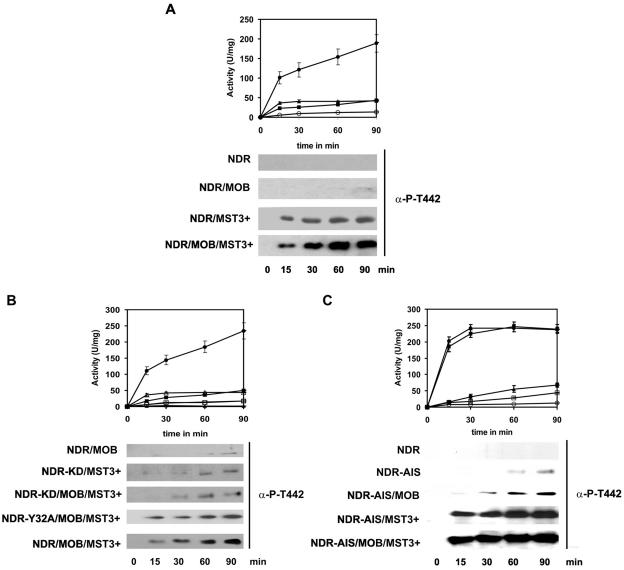
FIG. 2.
Analysis of NDR2 activation and hydrophobic motif phosphorylation by MST3 and MOB1A. (A) Activation and phosphorylation of wild-type NDR2. NDR2 SHP-NDR2 was incubated with GST-MOB1A (▪), GST-MST3 from OA-stimulated HEK293F cells (▴), both (•) or alone (○) under our standard NDR kinase assay conditions. (B) Activation and phosphorylation of kinase-dead and MOB-binding-deficient NDR2. SHP-NDR2 (○), SHP-NDR2/GST-MOB1A (▪), SHP-NDR2/MOB1A/MST3+ (•), SHP-NDR2K119A/MOB1A/MST3+ (▴), SHP-NDR2Y32A (□), and SHP-NDR2Y32A/MOB1A/MST3+ (Δ) were incubated under standard NDR kinase assay conditions. (C) Activation and phosphorylation of the NDR2-AIS mutant. SHP-NDR2 (○), SHP-NDR2-AIS (□), SHP-NDR2-AIS/GST-MOB1A (▴), SHP-NDR2-AIS/MST3+ (▪), and SHP-NDR2-AIS/GST-MOB1A/MST3+ (•) were incubated under standard NDR kinase assay conditions. At the time points indicated, NDR kinase activity was assayed with the NDR substrate peptide; results are expressed as specific activity. Results shown are means ± standard deviations of assays carried out in duplicate and are representative of two independent experiments. Error bars are only shown when larger than the size of the symbols. NDR hydrophobic motif phosphorylation was determined at each time point by Western blotting using the anti-P-T442 antibody.