FIG. 1.
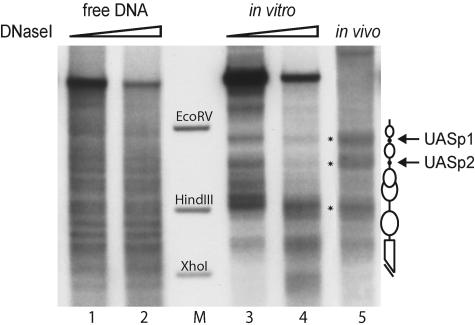
In vitro chromatin assembly with yeast extract generates the native chromatin structure at the PHO8 promoter. Limited DNase I digestion and secondary cleavage with BglII for indirect end labeling were performed with free DNA (lanes 1 and 2), chromatin assembled with yeast extract in vitro for 6 h (lanes 3 and 4), and wild-type yeast nuclei (lane 5). The marker bands correspond to the EcoRV-BglII, HindIII-BglII, and XhoI-BglII fragments of the PHO8 promoter (lane M). Schematics of the chromatin structure at the PHO8 promoter are on the right side of the gel. Ovals denote nucleosomes, black dots UASp elements, and the broken bar the open reading frame. Asterisks in the gel refer to the most distinguishing bands of the PHO8 promoter chromatin pattern. All samples were digested with a range of DNase I concentrations (ramps on top of the lanes). However, due to space limitations only representative lanes are shown.