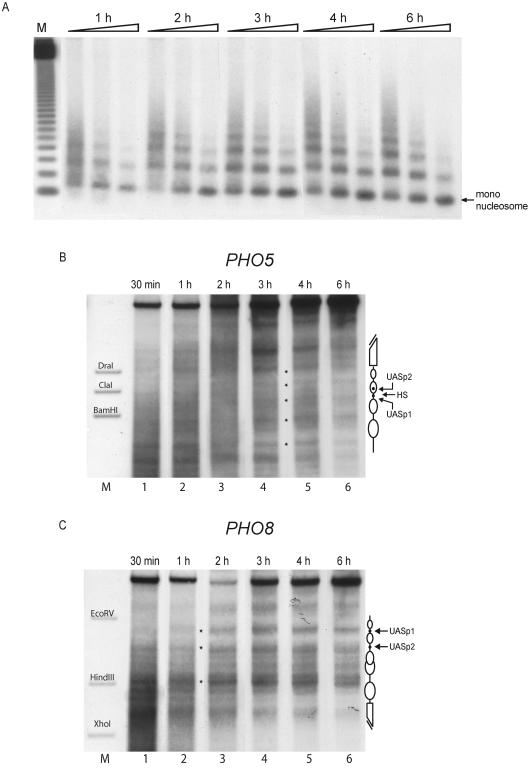
FIG. 2.
In a de novo in vitro assembly reaction nucleosomes become positioned more rapidly at the PHO8 promoter than at the PHO5 promoter. Assembly kinetics of a yeast extract in vitro assembly reaction with plasmids containing the PHO5 and PHO8 loci in the same reaction were monitored at the indicated time points by MNase digestion with specific probing for the PHO8 promoter region (A) and DNase I mapping probing for the PHO5 (B) or the PHO8 (C) promoter. Equivalent results as in panel A were also obtained by using a PHO5 promoter probe (not shown). Ramps on top of the lanes represent increasing MNase digestion times. Lane M (A) shows a 123-bp ladder (Gibco). The markers in panels B and C correspond to the ApaI-BamHI, ApaI-ClaI, and ApaI-DraI fragments of the PHO5 promoter and the EcoRV-BglII, HindIII-BglII, and XhoI-BglII fragments of the PHO8 promoter, respectively. Schematics of the chromatin structure at the PHO5 and PHO8 promoters are on the right side of the gels. Ovals denote nucleosomes, black dots UASp elements, and broken bars the open reading frame. HS in panel B denotes the linker region between nucleosomes −2 and −3 at the PHO5 promoter. Due to space limitations only representative lanes are shown.