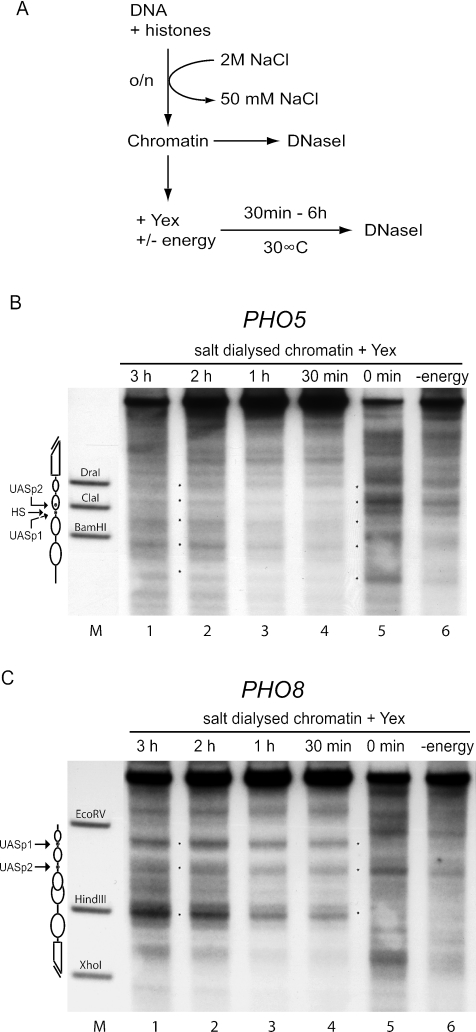
FIG. 5.
Nucleosomes preassembled by salt gradient dialysis become rapidly and properly repositioned after the addition of yeast extract. (A) Reaction scheme. Chromatin was generated by mixing DNA and histone octamers at a mass ratio of 1.1 and overnight (o/n) salt gradient dialysis from 2 M to 50 mM NaCl. The resulting chromatin was analyzed by DNase I mapping (DNase I) either right away or after an additional incubation with yeast extract (Yex) with or without an energy-regenerating system (energy) for up to 6 h at 30°C. (B and C) Chromatin after addition of yeast extract for the indicated times in the presence (lanes 1 to 5) or for 3 h in the absence (lane 6) of energy was analyzed by DNase I mapping with probing for the PHO5 (B) or the PHO8 (C) promoter. Marker lanes (M), schematics, and asterisks are as defined for Fig. 2. Due to space limitations only representative lanes are shown.