Abstract
Telomere length is controlled by a homeostatic mechanism that involves telomerase, telomere-associated proteins, and conventional replication machinery. Specifically, the coordinated actions of the lagging strand synthesis and telomerase have been argued. Although DNA polymerase α, an enzyme important for the lagging strand synthesis, has been indicated to function in telomere metabolism in yeasts and ciliates, it has not been characterized in higher eukaryotes. Here, we investigated the impact of compromised polymerase α activity on telomeres, using tsFT20 mouse mutant cells harboring a temperature-sensitive polymerase α mutant allele. When polymerase α was temperature-inducibly inactivated, we observed sequential events that included an initial extension of the G-tail followed by a marked increase in the overall telomere length occurring in telomerase-independent and -dependent manners, respectively. These alterations of telomeric DNA were accompanied by alterations of telomeric chromatin structures as revealed by quantitative chromatin immunoprecipitation and immunofluorescence analyses of TRF1 and POT1. Unexpectedly, polymerase α inhibition resulted in a significantly high incidence of Robertsonian chromosome fusions without noticeable increases in other types of chromosomal aberrations. These results indicate that although DNA polymerase α is essential for genome-wide DNA replication, hypomorphic activity leads to a rather specific spectrum of chromosomal abnormality.
Telomeres are specialized structures composed of telomeric DNA and associated proteins, and they protect the termini of linear chromosomes in eukaryotes. Telomeric DNA in most eukaryotic organisms consists of tandem arrays of a short repetitive sequence called a telomeric repeat, (TTAGGG) · (CCCTAA) in vertebrates. One strand of the telomeric repeat running towards the 3′ end of telomeres is G-rich (G-strand), whereas the other strand is C-rich (C-strand). The extreme end of the G-strand comprises a single-stranded DNA called the G-tail (66), which is produced by active processes involving C-strand degradation and/or passive processes involving the failure of the lagging strand synthesis to replicate the most terminal portion of telomeric DNA (the end replication problem) (39, 49, 65, 67).
Telomeres contain a variety of telomere-specific and nonspecific proteins, the most important of which are the proteins that are specifically bound to the telomeric double-stranded DNA, TRF1 and TRF2 in vertebrates (reviewed in reference 57). TRF proteins associate with telomeric repeats as homodimers through a specific DNA-protein interaction mediated by the C-terminal Myb-like DNA-binding domain. POT1 specifically recognizes the single-stranded G-tail via the OB fold domains at its N terminus (5, 31, 32). Recently, it has been proposed that six telomeric proteins, TRF1, TRF2, and POT1 as well as TIN2, Rap1, and TPP1 (originally known as PIP1, PTOP, or TINT1), comprise the core components of the mammalian telomeric complex (23, 34, 69).
Telomerase is a specialized reverse transcriptase that catalyzes the de novo synthesis of the G-tail using its intrinsic RNA template (33). In a given population of telomerase-positive cells, the telomere length is maintained within a relatively narrow range by means of a feedback mechanism operating between the telomere structure and the telomerase reaction. The accessibility of telomerase to its substrate (the 3′ terminus of the G-tail) is negatively regulated in cis by telomeric DNA-binding proteins (TRF1 in vertebrates) (41, 62). In budding yeast, the G-tail-binding protein Cdc13 recruits telomerase to the G-tail and activates it in a collaborative manner with Est1, a component of the yeast telomerase (15, 52, 59). The regulatory role of POT1 in telomerase appears to be more complex. Although several studies have suggested that POT1 is a positive regulator of telomerase (4, 10), subsequent studies have indicated that POT1 is a negative regulator of telomerase (36). Moreover, it was demonstrated that POT1-bound telomeric DNAs are not good substrates for telomerase in vitro (27). POT1 also participates in the protection of telomeric ends, because partial knockdown of POT1 led to reduced G-tail sizes and telomere deprotection as manifested by telomere fusions or the transient accumulation of DNA damage-responsive proteins, such as 53BP1 and γH2AX (22, 64, 68).
During S phase, telomeric DNA is replicated by a replication fork that originates and moves outward from a replication origin located somewhere in the inner regions of chromosomes. Accordingly, the G-strand is invariably replicated by the leading strand synthesis, whereas the C-strand is replicated by the lagging strand synthesis. While the internal regions of telomeres are replicated by the conventional DNA replication mechanism, several studies in different systems suggest that the telomerase reaction is intimately coupled with DNA replication. The telomerase reaction at artificially shortened telomeres appears to occur concomitantly with telomere replication in late S phase in budding yeast (40). In Oxytricha nova, telomeres and telomerase are colocalized at the replication band, a specialized structure undergoing replication (17). Because telomerase synthesizes only the G-strand, C-strand synthesis is required to complete the double-stranded telomeric DNA elongation. This task is believed to be performed by the lagging strand synthesis machinery. De novo addition of telomeric double-stranded DNA to an inducibly formed double-strand break did not occur in the absence of functional DNA polymerase α, polymerase δ, or primase, which are the enzymes required for the lagging strand synthesis (13). More importantly, the synthesis of the G-strand by telomerase did not occur on such occasions. Therefore, the lagging strand synthesis machinery is essential not only for the C-strand synthesis, but also for the G-strand synthesis by telomerase, suggesting highly coordinated actions of the two DNA-synthesizing apparatuses. Consistent with this notion, it was demonstrated that budding yeast polymerase α physically interacts with Cdc13 and that fission yeast polymerase α interacts with the telomerase catalytic subunit, Trt1 (12, 52).
Euplotes crassus provides a unique opportunity to study the role of the lagging strand synthesis independent of genome-wide replication. During the maturation of macronuclei, telomerase extensively adds telomeric DNAs to numerous newly formed DNA termini in the absence of general DNA replication. When cells were treated with aphidicolin, an inhibitor of DNA polymerases α, δ, and ɛ, during this specific period both G- and C-strand syntheses were perturbed, indicating that the lagging strand synthesis is closely coupled with the telomerase reaction (16). Indeed, one biochemical study has shown that a DNA primase component and telomerase reside in the same complexes during the developmental stage of macronuclei in this organism (53). Taken together, the lagging strand synthesis plays two roles in telomeric DNA synthesis: as a conventional DNA replication apparatus together with the leading strand synthesis and as a C-strand fill-in reaction coupled with the G-tail extension by telomerase (reviewed in reference 51).
The importance of the lagging strand synthesis machinery in telomere maintenance has been genetically demonstrated in budding yeast. It was observed that native telomere length was increased at the semipermissive temperature in yeast mutants of DNA polymerase α and Rfc1 that is involved in the lagging strand synthesis by loading PCNA (1, 7). Importantly, telomere elongation in DNA polymerase α mutants grown at the semipermissive temperature depends on the presence of active telomerase (1), reemphasizing the intimate relationship between the lagging strand synthesis apparatus and telomerase. The repression of telomere-inserted genes (telomere position effect [TPE]), which reflects telomeric heterochromatin, is also inactivated in DNA polymerase α mutant cells exposed to the semipermissive temperature, indicating that not only the DNA structure but also the chromatin structure at telomeres is altered by the compromised DNA polymerase α activity (2). Although such extensive studies in yeasts and ciliates have implicated the role of the lagging strand synthesis in telomere maintenance, similar conclusions have yet to be reached for higher eukaryotes.
tsFT20 cells, a mutant clone identified in N-methyl-N′-nitro-N-nitrosoguanidine-treated mouse mammary carcinoma FM3A cells, show temperature-sensitive growth: the cells grow as well as the parent FM3A cells at 33°C, but not at 39°C (44). The point mutation at codon 1180 of the DNA polymerase α p180-kD subunit gene leading to a Ser-to-Phe substitution is responsible for the phenotype as well as the heat-labile activity of DNA polymerase α in tsFT20 cells (26). In this study, we examined tsFT20 cells in order to investigate the role of DNA polymerase α in telomere regulation in mammalian cells.
MATERIALS AND METHODS
Plasmids.
cDNAs for mouse POT1, mouse TRF1, and the wild-type and the mutant forms of mouse DNA polymerase α p180 were expressed as fusion proteins with N-terminally inserted three hemagglutinin (HA) tags and three Flag tags using pMX-puro retrovirus vector (29) and the packaging cells Platinum E (PE) (43).
Cell culture and transfection.
tsFT20 cells were maintained at the permissive temperature of 33°C or the semipermissive temperature of 38°C in RPMI 1640 medium (Nissui) supplemented with 10% fetal bovine serum, 20 mM l-glutamine, NaHCO3, penicillin, streptomycin, and amphotericin. High-titer retroviral stocks were generated by transient transfection of PE packaging cells. Retroviral gene transfer was performed as previously described (54), and infected cells were selected by treatment with 10 μg/ml puromycin (Sigma).
Cell cycle analysis.
Subconfluent cultures were fixed with 70% ethanol and kept overnight at 4°C. Fixed cells were incubated with RNase (1 mg/ml; Sigma) for 30 min at 37°C, followed by staining with 50 μg/ml propidium iodide (Nacalai) for 30 min at room temperature. Flow cytometry was performed with a FACSCalibur (Becton Dickinson).
Hybridization of telomeric DNA.
Genomic DNA was digested with HinfI (0.3 U/μl; TaKaRa) overnight at 37°C. The digested DNA was fractionated on a 0.8% agarose gel in 1× Tris-acetate-EDTA. The gels were blotted onto Hybond-N+ membranes (Amersham), which were hybridized with the 5′-end-labeled (CCCTAA)4 oligonucleotide probe in hybridization buffer (50% formamide, 5× Denhardt's, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% sodium dodecyl sulfate [SDS]) overnight at 42°C. For in-gel hybridization, genomic DNA was treated with or without 0.25 U/μl exonuclease I (Exo I; New England Biolabs) overnight at 37°C. DNA was digested with HinfI overnight at 37°C and subjected to electrophoresis on a 0.8% agarose gel in 1× Tris-acetate-EDTA. In-gel hybridization analysis was performed with the 5′-end-labeled (CCCTAA)4 oligonucleotide probe in Church's hybridization buffer (7% SDS, 0.5 M NaPO4 [pH 7.2], 1 mM EDTA) overnight at 42°C (9). After obtaining autoradiographs, the gel was denatured with alkaline buffer (0.5 N NaOH and 150 mM NaCl) for 30 min, neutralized, and reprobed with the same probe to ensure that equal amounts of DNA were loaded in each lane.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed according to the methods described in the literature (24, 36). HA-POT1- and HA-TRF1-expressing tsFT20 cells were fixed with 1% formaldehyde in the medium for 10 min at 37°C, and this was followed by washing with phosphate-buffered saline (PBS; Nissui), lysis in cell lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl, pH 8.0), sonication, and centrifugation for 10 min at 4°C. For immunoprecipitation, anti-HA antibodies (16B12; Babco) and protein G-Sepharose beads (Amersham) were used. After washing the beads with NET buffer (0.5% NP-40, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, and 1 mM EGTA), coprecipitated DNA was extracted and suspended in Tris-EDTA. The samples were denatured with 0.4 N NaOH and blotted onto Hybond-N+ membranes. The membranes were hybridized with the 5′-end-labeled (CCCTAA)4 or (CA)12 oligonucleotide probe. The results were analyzed with a phosphorimager (Typhoon 9400; Amersham).
Telomerase knockdown by antisense oligonucleotides.
2′-O-MeRNA oligonucleotides were from GENSET OLIGOS. The sequences of the matched and mismatched anti-TR template oligonucleotides are 5′-CAGUUAGGGUUAG-3′ and 5′-CAGUUAGAAUUAG-3′, respectively, where the underlined nucleotides possess phosphorothioate linkages (21). A total of 2 × 105 tsFT20 cells in 2 ml of RPMI medium (without antibiotics) were transfected with 1.6 nmol 2′-O-MeRNA oligonucleotides using 8 μl of Lipofectamine 2000 (Invitrogen) in 1 ml of Opti-MEM l (Invitrogen). The cells were fed with fresh medium 3 days after the transfection. The following day, the cells were harvested and assayed for telomerase activity using the TeloChaser kit (TOYOBO) based on the stretch PCR method (60).
Immunofluorescence (IF).
Cells plated on cover slides were prepermeabilized in 0.1% Triton buffer (0.1% Triton X-100, 20 mM HEPES-KOH [pH 7.4], 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) on ice for 5 min and fixed with 2% formaldehyde in PBS on ice for 10 min. They were washed twice with PBS and treated with Triton buffer (same as 0.1% Triton buffer except 0.5% Triton X-100 was used) at room temperature for 10 min. Subsequent steps were performed as previously reported (45). Images were recorded with a Delta Vision restoration microscope (Applied Precision) equipped with a 60× objective lens. 4,6-Diamidino-2-phenylindole (DAPI), Alexa 488, and cyanine 3 (Cy3) were excited by lights at 360 nm (±20 nm), 490 nm (±10 nm), and 555 nm (±14 nm), respectively, and images were collected at 457 nm (±25 nm), 528 nm (±14 nm), and 617 nm (±37 nm), respectively. Rabbit anti-mouse TRF1 antibodies (25) were a kind gift from Y. Shinkai (Kyoto University Institute for Virus Research).
Quantitative image analysis.
Alexa 488 and Cy3 signals that represented TRF1 and HA-POT1, respectively, were processed by the deconvolution software in Delta Vision and Graphic Converter (Lemke Software), and signal intensity was recorded for each pixel. One z-section was chosen for each scanned field. The background signal was determined so that the telomeric signals appeared as punctuate spots. Once a threshold value was determined, it was applied to the sections examined in the same scanned field. Signals above the threshold value were extracted, and positive pixels continuous in the x-, y-, or z-section were merged to form signal particles. The diameters of all the particles were less than 0.4 μm. In a previous similar study, single telomeres were detected as fluorescence in situ hybridization (FISH) signals showing a similar size distribution, suggesting that most, if not all, TRF1 and POT1 signals in our study represented single telomeres (50). The signal intensities (after subtracting the background) were integrated for each particle, and the value was considered to represent the intensity of the cognate telomeric fluorescence signal. Histograms of the signal intensity distribution were created using Kaleida Graph (Synergy Software). The difference between distributions obtained from different samples was statistically analyzed by the Wilcoxon-Mann-Whitney test.
FISH.
Cells were grown for 4 to 12 h in the presence of 0.1 μg/ml Colcemid. After treatment with hypotonic buffer (10 mM Tris HCl, 10 mM NaCl, and 5 mM MgCl2) for 30 min, the cells were fixed with methanol-acetic acid, dropped on glass slides, and dried. Telomere FISH was performed essentially as described in reference 72. The glass slides were washed with PBS and fixed with 3% formaldehyde for 10 min. A hybridization mixture containing 0.3 μg/ml Cy3-conjugated (CCCTAA)3 PNA probe (Sawady) in 50% formamide, 2× SSC, 10% dextran sulfate, and 5× Denhardt's solution was prepared. The hybridization mixture was preheated at 80°C for 3 min and incubated with the glass slides overnight at room temperature. The glass slides were washed with 2× SSC and 50% formamide twice, followed by washing with 2× SSC twice and staining with 1 μg/ml DAPI. Cells were analyzed with a Delta Vision restoration microscope (Applied Precision).
RESULTS
Telomeres are elongated in tsFT20 cells at semipermissive temperature.
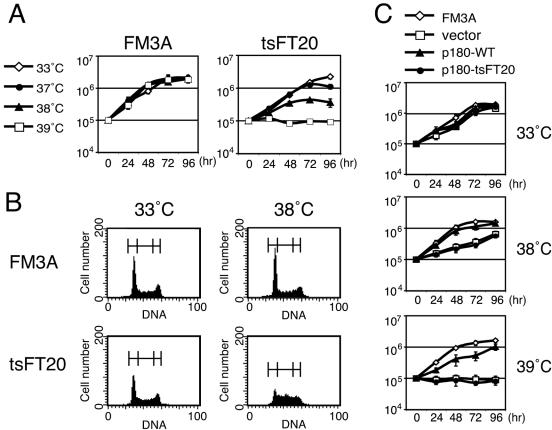
It has been reported that tsFT20 cells grow as well as the parent FM3A cells at the permissive temperature of 33°C, but not at the nonpermissive temperature of 39°C (44); this was recapitulated in this study (Fig. 1A). We cultured the cells at various temperatures and found that they grew slowly at 38°C. The doubling times of tsFT20 cells at 33°C and 38°C were 24 h and 30 h, respectively, whereas those of FM3A cells at 33°C and 38°C were from 16 to 18 h. tsFT20 cells cultured at 38°C exhibited an increase in the fraction of cells in S phase compared with those cultured at 33°C (53.8% versus 36.5%). In contrast, FM3A cells cultured at these temperatures showed essentially the same fractions of cells in S phase (33.5% and 29.3% at 33°C and 38°C, respectively) (Fig. 1B). tsFT20 cells cultured at 38°C showed reduced levels of bromodeoxyuridine incorporation (data not shown). These results are consistent with the idea that DNA polymerase α activity in tsFT20 cells at 38°C is partially inactivated. The slow growth (Fig. 1C) and the increase of the S-phase fraction (data not shown) of tsFT20 cells at 38°C were rescued when the cells were transfected with retrovirus encoding the wild-type p180 subunit (p180-WT) cDNA, but not with those encoding the tsFT20-derived temperature-sensitive mutant allele (p180-tsFT20) cDNA or the empty vector (Fig. 1C). Taken together, we concluded that the mutant DNA polymerase α in tsFT20 cells has a semipermissive temperature of 38°C, which was used in this study to compromise DNA polymerase α activity in a temperature-inducible manner.
FIG. 1.
Temperature-dependent growth of tsFT20 cells. A. Growth curves of FM3A (wild type) and tsFT20 mutant at 33°C, 37°C, 38°C, and 39°C. tsFT20 exhibited significant yet retarded growth at 38°C and no growth at 39°C. Cell density was adjusted to 105/ml at 0 h, and the cultures were continued for 96 h. Cell density was measured at intervals as indicated. B. Cell cycle analysis of FM3A and tsFT20 cells showing logarithmic growth at 33°C and 38°C by FACScan. tsFT20 cells cultured at 38°C showed an increase in the fraction of cells in S phase. The percentages of cells in G1, S, and G2+M phases (indicated by brackets above the peak curves) were as follows: 43.2%, 33.5%, and 23.3% for FM3A at 33°C; 49.5%, 29.3%, and 21.2% for FM3A at 38°C; 44.6%, 36.5%, and 18.9% for tsFT20 at 33°C; and 28.7%, 53.8%, and 17.5% for tsFT20 at 38°C. C. tsFT20 cells expressing the wild-type p180 subunit of DNA polymerase α bypassed the slow growth at high temperatures. tsFT20 cells were infected with retroviruses encoding the indicated proteins, and the infected cells were selected by treatment with puromycin for 5 days. The selected cells as well as FM3A cells were used for obtaining growth curves as described for panel A. Mock-infected, vector; wild-type p180 subunit of DNA polymerase α, p180-WT; temperature-sensitive mutant form of p180 derived from tsFT20,p180-tsFT20.
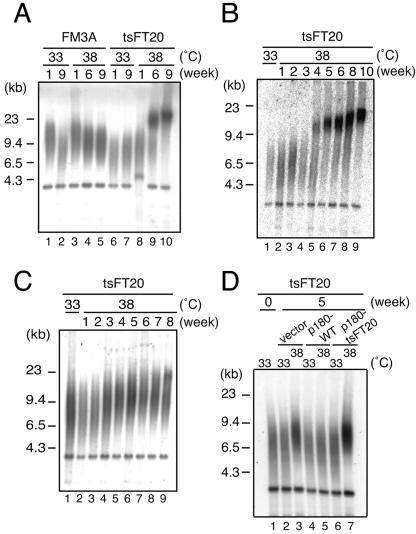
We measured the telomere lengths of FM3A and tsFT20 cells cultured at permissive (33°C) and semipermissive (38°C) temperatures. Both cell types were routinely maintained at 33°C and divided into two populations, one kept at 33°C and the other shifted to 38°C. Genomic DNA was isolated, digested with HinfI, and analyzed by Southern hybridization using the vertebrate telomeric oligonucleotide probe, (CCCTAA)4. Mus musculus typically shows long telomeric repeats ranging from approximately 50 to 100 kb (28). However, FM3A cells possessed relatively short (6 to 9 kb) telomeric repeats in all cases of cultures at 33°C or 38°C for 1, 6, or 9 weeks (Fig. 2A, lanes 1 to 5). The band at ∼4 kb is derived from internal telomeric repeats. tsFT20 cells grown at 33°C for 1 or 9 weeks exhibited telomeric repeats within the same length range (lanes 6 and 7). In contrast, tsFT20 cells kept at 38°C showed progressive and marked elongation of telomeric repeats during the 9-week culture. The repeat length did not noticeably change after incubation for 1 week at the semipermissive temperature (lane 8) but was markedly increased after a 6-week incubation (lane 9). At this time, two telomeric signals were apparent, a smear signal around 7 to 9 kb and the other around 20 to 30 kb. After a 9-week culture, the 20- to 30-kb signal increased in size further, and the smear signal appeared to be larger than the 20- to 30-kb signal, suggesting that the telomeric repeat growth continued during the incubation period of 6 to 9 weeks at 38°C (lane 10).
FIG. 2.
Elongation of telomeric repeats in tsFT20 at semipermissive temperature. HinfI-digested genomic DNAs isolated from FM3A or tsFT20 cells cultured at 33°C or 38°C for the indicated periods were analyzed for telomeric repeats by Southern hybridization using the telomeric repeat probe (CCCTAA)4. A. tsFT20 exhibited gradual elongation of telomeric repeats during culture at 38°C for 9 weeks. B. Some tsFT20 populations displayed sudden onset of telomeric repeat elongation. C. Some tsFT20 populations displayed gradual telomeric repeat elongation. D. Telomeric repeat elongation does not occur in tsFT20 cells rescued by p180-WT. tsFT20 cells expressing the indicated protein were measured for telomeric repeat length after incubation at 33°C or 38°C for 5 weeks. tsFT20 cells were infected with the control retrovirus vector (vector), the retrovirus encoding the wild-type p180 (p180-WT), or the temperature-sensitive p180 mutant derived from tsFT20 (p180-tsFT20), selected by puromycin treatment, and analyzed.
By performing the experiments several times independently and measuring the elongation kinetics of the telomeric repeat, we noticed two apparently distinct elongation patterns. In some culture populations, it appeared that the telomeric repeats grew abruptly within a relatively short window of time. In one case, shown in Fig. 2B, a sudden onset of elongation was observed between the 3- and 4-week time points after the temperature shift. The size and intensity of the major signal kept increasing thereafter, indicating that telomere elongation continued (lanes 5 to 9). In other culture populations, as exemplified in Fig. 2C, the elongation rate of telomeric repeats appeared to be relatively constant, and the end point after a 9-week culture at 38°C was a relatively mild telomeric extension compared with cases that showed sudden elongation. In a total of eight independent cultures, the type of elongations shown in Fig. 2B and 2C were observed five and three times, respectively. Interestingly, any given culture population displayed either type of telomere elongation with some exceptions (see below), and the time course of elongation was relatively reproducible within one type (see Fig. S1 in the supplemental material). We do not know at this moment what is responsible for the difference in behavior of telomeres in response to the temperature shift, and it appears to be determined stochastically. We also noted that in some cases, as shown in Fig. 2A, the population was a mixture of two subpopulations that showed two distinct elongation patterns, judging from the presence of the suddenly elongating telomere populations and the gradually elongating ones. We do not know whether different cell populations in the culture or different telomeres in one cell behave differently in such cases. We concluded that the telomeric repeat length of tsFT20 cells is markedly and continuously increased after the temperature is shifted from 33°C to 38°C.
To confirm that the telomere elongation in tsFT20 cells at 38°C is due to the defect of the p180 subunit of DNA polymerase α, a complementation experiment was performed using tsFT20 cells expressing p180-WT or p180-tsFT20 (described in Fig. 1C). These cells were maintained at 33°C or 38°C for 5 weeks, and their telomere lengths were analyzed by Southern hybridization (Fig. 2D). We found that tsFT20 cells rescued by p180-WT did not show telomere elongation at 38°C (lane 5). On the other hand, tsFT20 cells expressing p180-tsFT20 or infected with the control vector showed elongated telomeres at 38°C (lanes 3 and 7). These results indicate that telomere elongation in the tsFT20 mutant is caused by the specific defect of DNA polymerase α at 38°C.
G-tail extension prior to telomeric double-stranded DNA elongation.
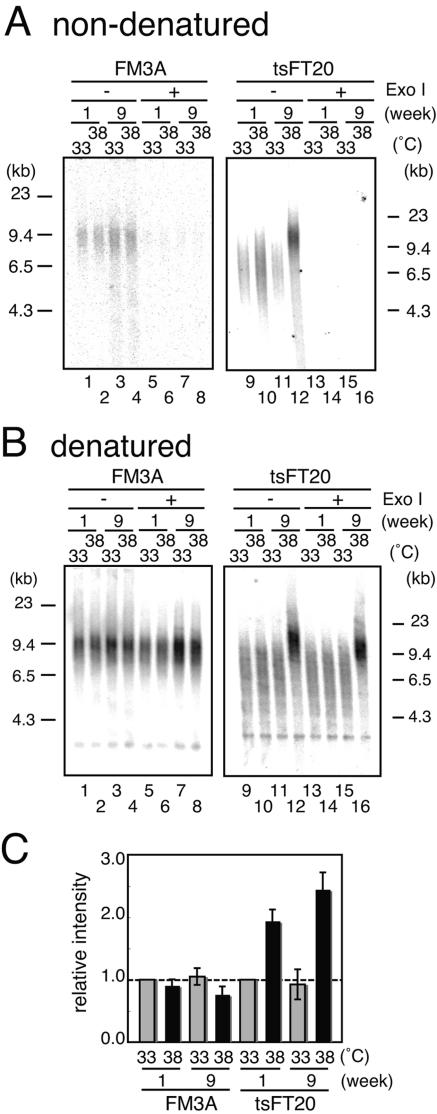
To explain telomere elongation in tsFT20 cells at the semipermissive temperature, we reasoned that the defective DNA polymerase α produced abnormal telomeric DNA structures. Because DNA polymerase α is mainly involved in the lagging strand synthesis, its mutant may show a defect of the telomeric C-strand synthesis that leads to the extension of the G-tail. The amount of the G-tail was measured using the nondenaturing in-gel hybridization technique for FM3A and tsFT20 cells grown at 33°C or 38°C. HinfI-digested DNA run in a gel was directly hybridized with the (CCCTAA)4 oligonucleotide probe under neutral conditions and subjected to phosphorimaging analysis. The gel was subsequently denatured and rehybridized with the (CCCTAA)4 oligonucleotide probe, and autoradiographs were obtained (Fig. 3). The signals obtained by the nondenaturing in-gel hybridization were sensitive to the treatment with Exo I, which digests single-stranded DNA from the 3′ termini prior to the HinfI digestion, suggesting that they were indeed derived from the G-tail (Fig. 3A, lanes 5 to 8 and 13 to 16). Signals detected after rehybridization of the denatured gel (Fig. 3B) showed similar intensities except for those of tsFT20 cells grown at 38°C for 9 weeks, which showed extensive telomeric elongation that excluded a direct comparison with the other lanes. However, the ∼4-kb band derived from internally located telomeric repeat sequences was detected at the same intensity for all cases. The results together indicated that the same amounts of genomic DNAs that were completely digested were analyzed in Fig. 3A.
FIG. 3.
tsFT20 at 38°C showed an extended G-tail before telomeric DNA elongated. A. G-tail signals. DNA was prepared from FM3A or tsFT20 cells that were cultured at 33°C or 38°C for 1 or 9 weeks, treated or untreated with Exo I, and HinfI digested. After electrophoresis, the DNA in the gel was hybridized with the (CCCTAA)4 probe under nondenaturing conditions. The gel was exposed to a film to obtain an autoradiograph. B. The gel used in panel A was denatured and rehybridized with the (CCCTAA)4 probe. C. The intensities obtained in panel A were normalized to the intensity of the ∼4-kb signal in panel B, and the relative values are shown such that the value for FM3A or tsFT20 cells cultured at 33°C for 1 week is set as 1.
FM3A cells showed similar signal intensities under nondenaturing conditions for all cases when grown at 33°C or 38°C for 1 or 9 weeks (Fig. 3A, lanes 1 to 4), demonstrating that the G-tail length did not change much despite the different culture conditions. In contrast, tsFT20 cells cultured at 38°C showed significant increases of the signal intensity in the nondenatured gel, compared with the control tsFT20 cells at 33°C (Fig. 3A, lanes 9 to 12). tsFT20 cells grown at 38°C for 1 week showed a signal with increased intensity (Fig. 3C) (1.9-fold ± 0.18-fold; n = 3) (the G-tail signal intensity is normalized to the loading control, the intensity of the ∼4-kb band in Fig. 3B) and the same mobility compared with those grown at 33°C (Fig. 3A, lanes 9 and 10). These results are consistent with the observation that the telomeric sequences did not elongate at this stage (Fig. 3B, lane 10) and suggest that the G-tail had already extended prior to the elongation of the telomeric repeats. After a 9-week culture at 38°C, tsFT20 cells displayed elongated telomeres (Fig. 3B, lane 12) and the G-tail showed a further increase in signal intensity (Fig. 3C) (2.4-fold ± 0.27-fold; n = 3) and consistently retarded mobility (Fig. 3A, lane 12). The G-tail extension observed in tsFT20 cells at 38°C was caused by compromised DNA polymerase α activity, because tsFT20 cells expressing p180-WT did not show this phenotype, whereas those expressing p180-tsFT20 did (data not shown). We concluded that the alteration of telomeric DNA occurs in a two-step manner in DNA polymerase α-defective tsFT20 cells: first, the length of the single-stranded region of telomeres is increased, and this is followed by the elongation of the double-stranded telomeric repeats.
Ordered increases of POT1 and TRF1 bound to telomeres in tsFT20 cells at semipermissive temperature.
Such an ordered sequence of G-tail extension and telomeric DNA elongation was previously observed in a temperature-sensitive yeast DNA polymerase α mutant (2). It was also reported that TPE is inactivated in the mutant at the semipermissive temperature, suggesting that not only the structure of telomeric DNA but also that of telomeric chromatin was changed by defective DNA polymerase α (2). To examine whether telomeric chromatin is also altered in tsFT20 cells cultured at 38°C, we focused on two telomeric DNA-binding proteins, TRF1 and POT1, which bind to double-stranded and single-stranded telomeric DNA, respectively (5, 8). Using retrovirus vectors, we generated tsFT20 cells that stably expressed N-terminally HA-tagged mouse POT1 or mouse TRF1 (tsFT20-HA-POT1 or tsFT20-HA-TRF1, respectively). Indirect IF experiments using anti-HA antibodies indicated that HA-POT1 and HA-TRF1 colocalized with endogenous TRF1 detected with anti-TRF1 antibodies (data not shown). Telomere lengths were not significantly altered by the expression of HA-POT1 or HA-TRF1 (Fig. 4A, lanes 1 to 3 and lanes 7 to 9).
FIG. 4.
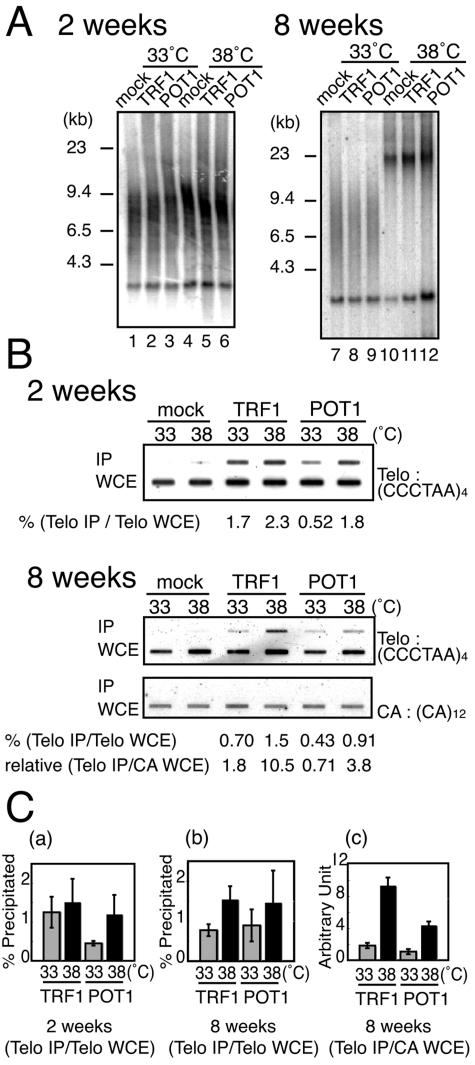
ChIP analyses revealed that POT1 and TRF1 sequentially accumulate at telomeres. A. tsFT20 cells stably expressing the HA-tagged indicated protein were obtained by infecting the cells with retrovirus expression vectors, followed by puromycin selection. The cells were cultured at 33°C or 38°C for 2 or 8 weeks, and DNAs were digested with HinfI and subjected to Southern hybridization using the telomeric (CCCTAA)4 probe. B. Telomeric DNA present in anti-HA immunoprecipitates from tsFT20 cells expressing HA-TRF1 or HA-POT1. tsFT20-HA-POT1, tsFT20-HA-TRF1, and mock-infected tsFT20 cells were cultured at 33°C or 38°C as indicated and subjected to ChIP assay. DNAs present in the immunoprecipitates obtained with anti-HA antibodies were directly hybridized with indicated probes. The applied amount of WCE samples was equivalent to 1/16 of that of IP samples. The fraction of telomeric DNA specifically precipitated among that in WCE (Telo IP/Telo WCE) was calculated as follows: [(telomeric DNA signal in precipitate)/(telomeric DNA signal in WCE)](TRF1 or POT1) − [(telomeric DNA signal in precipitate)/(telomeric DNA signal in WCE)](mock). For the cells incubated at 38°C for 8 weeks, the relative amount of telomeric DNA specifically precipitated per unit cell (Telo IP/CA WCE) was calculated as [(telomeric DNA signal in precipitate)/(CA signal in WCE)](TRF1 or POT1) − [(telomeric DNA signal in precipitate)/(CA signal in WCE)](mock). These values are expressed in arbitrary units. C. Quantitation of ChIP results. The results obtained from three independent experiments including that shown in B were quantitated. (a and b) Fraction (percentage) of precipitated telomeric DNAs among the total telomeric DNAs for cells incubated for 2 weeks (a) and 8 weeks (b) at the indicated temperatures. (c) Amount of precipitated telomeric DNAs on a unit cell basis (expressed in an arbitrary unit) for cells incubated for 8 weeks at the indicated temperatures.
To analyze the amounts of POT1 and TRF1 that bind to telomeres during the two-step alteration of telomeric DNA, we incubated tsFT20-HA-POT1 and tsFT20-HA-TRF1, as well as mock-infected tsFT20, at 38°C for 2 or 8 weeks. The expression levels of HA-POT1 in tsFT20-HA-POT1 and HA-TRF1 in tsFT20-HA-TRF1 did not change regardless of culture at 33°C or 38°C (data not shown). The cells cultured at 38°C for 2 weeks did not show noticeable double-stranded telomeric DNA elongation compared with those cultured at 33°C, demonstrating that the cells were at the first step of telomeric DNA alteration (Fig. 4A, 2 weeks). The cells grown at 38°C for 8 weeks displayed extensive telomeric DNA elongation, indicating that they had entered the second step of telomeric DNA alteration (Fig. 4A, weeks). These cell samples were analyzed for the presence of telomeric DNA-bound proteins by the ChIP assay (Fig. 4B). HA-tagged proteins were immunoprecipitated from formalin-cross-linked cell samples using anti-HA antibodies. The presence of telomeric DNA in the de-cross-linked immunoprecipitates was detected by slot blot hybridization with the telomeric (CCCTAA)4 oligonucleotide probe without any amplification step. Little, if any, telomeric DNA was detected in the immunoprecipitates from mock-infected tsFT20 cells cultured at 33°C or 38°C for 2 or 8 weeks (Fig. 4B). As a second negative control, the (CA)12 oligonucleotide probe did not detect any signals in the immunoprecipitates of either case (Fig. 4B, 8-week samples). These results together demonstrated the specificity of the ChIP assay. The hybridization was quantitative, because when we loaded serially diluted samples, we found that the signal intensities were changed in a linear fashion (data not shown).
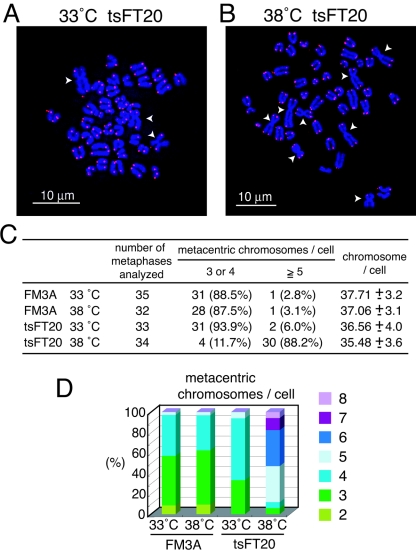
FIG. 8.
Robertsonian fusions in tsFT20 cells. A and B. FISH analyses of metaphase chromosome spreads prepared from tsFT20 cells cultured at 33°C (A) and 38°C (B) for 6 weeks. DNAs stained with DAPI are shown in blue, and telomeric repeats revealed by the Cy3-labeled telomeric repeat PNA probe are shown in red. White arrowheads indicate metacentric chromosomes formed by the Robertsonian fusion. Note that no detectable telomeric repeat signal is present at the fusion points. C. Metaphase spreads obtained from indicated cells were scored according to the number of Rb fusions and the number of total chromosomes. D. Frequencies of spreads displaying different numbers of Rb fusions.
We calculated the fraction of telomeric DNAs immunoprecipitated by anti-HA antibodies relative to the total telomeric DNAs in the whole-cell extract (WCE). Approximately 0.4 to 2.4% of telomeric DNAs present in WCE (after subtracting the background precipitates deduced by the mock expressing cells) were immunoprecipitated in our system. In a previous study, approximately 20 to 30% of total telomeric DNAs were consistently detected in anti-TRF1 immunoprecipitates among different cell lines showing different telomere lengths, suggesting that TRF1 associates with telomeres in a stoichiometric manner with telomeric DNA length (36). The reason for the relatively low recovery of the telomeric DNA in the immunoprecipitates in this study compared with those in the previous study is not known.
In the case of 2-week cultures, significantly larger amounts of telomeric DNAs were found in anti-HA-POT1 precipitates from cells incubated at 38°C than from cells incubated at 33°C (Fig. 4C, graph a; n = 3), concomitantly with the elongation of single-stranded G-tail in tsFT20 at 38°C. In contrast, those amounts present in anti-HA-TRF1 precipitates were within the same range between cells cultured at 33°C or 38°C (n = 3), a result consistent with the observation that the double-stranded telomeric DNA did not elongate at this stage.
When we analyzed the ChIP results obtained for cells in the second step of telomeric changes (8-week incubation), it was necessary to take into account the telomere elongation. For the normalization, we used the (CA)12 signal in WCE as the total DNA of unit cells. Total telomeric DNA per unit cell was increased by approximately twofold in tsFT20-HA-TRF1 and tsFT20-HA-POT1 cells cultured at 38°C, compared with that in cells cultured at 33°C for 8 weeks (2.2-fold ± 0.74-fold, n = 3, and 2.0-fold ± 0.40-fold, n = 3, respectively). Relative telomeric DNA per unit cell was calculated from the following equation: [(CCCTAA)4 signal in WCE]/[(CA)12 signal in WCE]. The relative amount of precipitated telomeric DNA per unit cell (i.e., per single telomere locus) was calculated from the equation [(CCCTAA)4 signal in IP]/[(CA)12 signal in WCE], and the results indicated that significantly larger amounts of telomeric DNAs were present in both HA-TRF1 and HA-POT1 precipitates from cells incubated at 38°C than in those from cells incubated at 33°C (5.1-fold, n = 3, and 4.1-fold, n = 3, respectively) (Fig. 4C, graph c).
It was also suggested that the relative density of TRF1 and POT1 at telomeres {the amount of telomeric DNA per unit telomeric DNA length, calculated from the equation [(CCCTAA)4 signal in IP]/[(CCCTAA)4 signal in WCE]}, was increased in these cells, although the increase was not statistically significant for POT1 (2.1-fold, n = 3, and 1.7-fold, n = 3, respectively) (Fig. 4C, graph b). Taken together, it was found that the absolute amounts of, and possibly the relative densities of, HA-POT1 at telomeres were increased in tsFT20-HA-POT1 cells incubated at 38°C for 2 weeks and thereafter. In contrast, increases in the abundance of HA-TRF1 were not observed until the second step of telomeric changes (8-week incubation at 38°C).
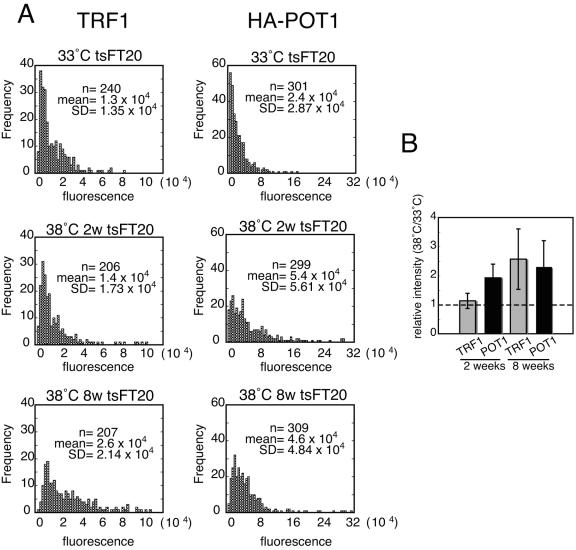
As a second means to quantitate the relative abundance of POT1 and TRF at telomeres, we measured the relative signal intensity of HA-POT1 and the endogenous TRF1 immunofluorescence in cells detected with anti-HA and anti-TRF1 antibodies, respectively. tsFT20-HA-POT1 cells were prepermeabilized to remove soluble proteins, fixed, and reacted with antibodies. Signal intensities were recorded for each pixel in one nucleus, and pixels showing signals above the background level were extracted (see Materials and Methods). The distribution of particle signal intensity for each cell culture (200 to 300 segments for each condition) was analyzed (Fig. 5A). The Wilcoxon-Mann-Whitney test indicated that the median intensities of the TRF1 fluorescence signals were within the same range between cells cultured at 33°C and 38°C for 2 weeks (P = 0.58). In contrast, the median intensities of the TRF1 fluorescence signals in cells cultured at 38°C for 8 weeks, and also the HA-POT1 fluorescence signals in cells cultured at 38°C for 2 or 8 weeks, were significantly higher than those in cells cultured at 33°C (P < 0.001 in all cases). These results indicated that telomere-bound HA-POT1 but not TRF1 was increased in tsFT20-HA-POT1 cells after a 2-week incubation at 38°C, and both HA-POT1 and TRF1 were increased after an 8-week incubation compared with those cultured at 33°C.
FIG. 5.
Higher signal intensities of TRF1 and HA-POT1 in tsFT20-HA-POT1 cells cultured at semipermissive temperature than those in cells at permissive temperature. tsFT20-HA-POT1 cells were simultaneously stained for HA-POT1 and endogenous TRF1 by IF using anti-HA and anti-TRF1 antibodies, respectively. HA-POT1 and TRF1 signals at telomeres were extracted and quantitated by the method described in Materials and Methods. A. Frequency distributions of telomeric HA-POT1 and TRF1 fluorescence signal intensities in tsFT20-HA-POT1 cells cultured under the indicated conditions. The frequency distributions of HA-POT1 and TRF1 in all the cell cultures at 38°C, except that of TRF1 in cells at 38°C for 2 weeks, were significantly different from those of cells cultured at 33°C (P < 0.001, Wilcoxon-Mann-Whitney test). The number of analyzed telomere signals (n), the mean value (mean), and the standard deviation (SD) of the signal intensities are shown. B. The RMSI38/33 values for TRF1 and HA-POT1 in cells cultured for 2 or 8 weeks were calculated from four independent experiments.
The mean signal intensity was calculated for all particles in cells under one growth condition after subtracting the background signal. The relative mean signal intensity (RMSI38/33) was obtained by dividing the value for cells cultured at 38°C by that for cells cultured at 33°C. Experiments were performed independently four times, and the obtained RMSI38/33 values for TRF1 for cells incubated for 2 weeks and 8 weeks were 1.13-fold ± 0.26-fold and 2.6-fold ± 1.05-fold, respectively. In contrast, the RMSI38/33 values for HA-POT1 after 2- and 8-week incubations were 1.9-fold ± 0.47-fold and 2.3-fold ± 0.90-fold, respectively (Fig. 5B). Because the “background level” varied to some extent from sample to sample, the absolute values obtained in this analysis should be considered semiquantitative. However, it is noteworthy that the RMSI38/33 values obtained in these experiments were consistent with those obtained in the ChIP experiments (Fig. 4).
Taken together, these experiments suggest that impaired DNA polymerase α activity results in dynamic changes of not only telomeric DNA but also telomeric chromatin structures. In the initial step of the changes (2 weeks at 38°C), POT1 rather than TRF1 associates with telomeric DNA in higher absolute amounts than in cells cultured at 33°C, concomitant with possessing longer G-tails without overall telomeric DNA elongation. In the second stage (8 weeks at 38°C), larger amounts of both TRF1 and POT1 associate with telomeric DNA, concomitant with possessing long telomeric DNA and G-tails. Therefore, it appears that the amounts of POT1 and TRF1 present at telomeres are largely correlated with the lengths of single-stranded and double-stranded telomeric regions, respectively, in this system.
Telomerase is required for telomeric repeat elongation, but not for G-tail extension, in tsFT20 at semipermissive temperature.
The G-tail extension and the telomeric repeat elongation may have several possible mechanisms. The G-tail may be exposed simply because of the failure of the C-strand synthesis, or it may be elongated by a net synthesis, for example, by telomerase. Similarly, the telomeric repeat may be synthesized by telomerase or such mechanisms as those involving DNA recombination. To investigate the role of telomerase in the two-step alteration of telomeric DNA in tsFT20 cells at the semipermissive temperature, we knocked down the telomerase activity using a 13-nucleotide phosphorothioate oligonucleotide that targets telomerase RNA (TR) (21). The anti-TR oligonucleotide contains the sequence 5′-AGGGUUAG-3′, which is completely complementary to the template region of mouse telomerase RNA (mTR), 5′-CUAACCCU-3′. As control, we also tested an anti-TR-Cr oligonucleotide that contains the sequence 5′-AGAAUUAG-3′, having two nucleotides not complementary to mTR (underlined). These two oligonucleotides were transfected into tsFT20 cells cultured at 33°C or 38°C separately, and telomerase activity was measured using the stretch PCR assay 4 days after the transfection (60). Telomerase was significantly inhibited not by anti-TR-Cr, but by anti-TR, at 33°C and 38°C (Fig. 6B, lanes 4 to 7). The activity was sensitive to RNase pretreatment, suggesting that the product was indeed derived from telomerase activity (lanes 8 to 11). Then, tsFT20 cells were transfected with the oligonucleotides every 3 days during a 5-week culture. tsFT20 cells transfected with anti-TR or anti-TR-Cr for 5 weeks at 33°C showed no change in telomere length compared with those observed prior to the transfection, indicating that the oligonucleotide transfection procedure itself does not affect telomere length (Fig. 6C, lanes 1 to 3). Interestingly, telomere length was not increased at 38°C when telomerase was inhibited by transfecting anti-TR during the 5-week culture, whereas the same treatment with anti-TR-Cr had no effect on the telomere elongation (lanes 4 and 5). These results indicate that telomerase activity is required for telomeric repeat elongation in tsFT20 cells at 38°C.
FIG. 6.
Telomerase activity is required for telomeric repeat elongation, but not for G-tail extension. A. Anti-TR, which is complementary to the template region of hTR (human telomerase RNA) and mTR, and the control oligonucleotide (anti-TR-Cr), which possesses two mismatched nucleotides in the template region. B. tsFT20 cells cultured at 33°C or 38°C were transfected with anti-TR or anti-TR-Cr oligonucleotides and harvested 4 days later. Then, the cells were analyzed for telomerase activity according to the methods described in reference 60. C. tsFT20 cells cultured at 33°C or 38°C were transfected with anti-TR or anti-TR-Cr oligonucleotides at 3-day intervals for 5 weeks. Telomeric repeat length was measured as described in the legend for Fig. 2. Similar results were obtained from three independent experiments. D. tsFT20 cells cultured at 33°C or 38°C were transfected with anti-TR or anti-TR-Cr oligonucleotides at 3-day intervals for 2 weeks. G-tail length was measured as described for Fig. 3. Similar results were obtained from two independent experiments. E. The G-tail signal intensities obtained in panel D were quantitated. Values obtained for cells grown at 33°C are set at 1.0.
We next investigated whether telomerase is also required for the G-tail extension observed during the initial step of telomeric DNA alteration in tsFT20 cells cultured at 38°C. tsFT20 cells cultured at 33°C or 38°C were transfected with either anti-TR or anti-TR-Cr every 3 days and harvested 2 weeks after the temperature shift. The G-tail was examined by the in-gel hybridization technique as described above. tsFT20 cells cultured at 38°C for 2 weeks showed increased G-tail signal intensity as detected by the (CCCTAA)4 probe irrespective of the transfection of anti-TR or anti-TR-Cr, compared with cells cultured at 33°C (Fig. 6D, lanes 1 to 4). These signals were sensitive to the pretreatment with Exo I, demonstrating that they were indeed derived from the 3′-terminal fragment (lanes 5 to 8). The signals obtained under denaturing conditions confirmed that the same amount of DNA that was completely digested was loaded to each lane (lanes 9 to 16). When the G-tail signal was quantitated, we observed similar levels (1.6- to 1.8-fold) of increases in both cases of anti-TR- and anti-TR-Cr-transfected cells at 38°C compared with those at 33°C. Together, we concluded that telomerase activity is not required for the G-tail extension in the initial step of telomeric DNA alteration in tsFT20 cells cultured at 38°C. This conclusion suggests that the G-tail extension is not due to the net synthesis of the G-strand.
tsFT20 cells that experience telomere elongation adapt to new telomere length setting.
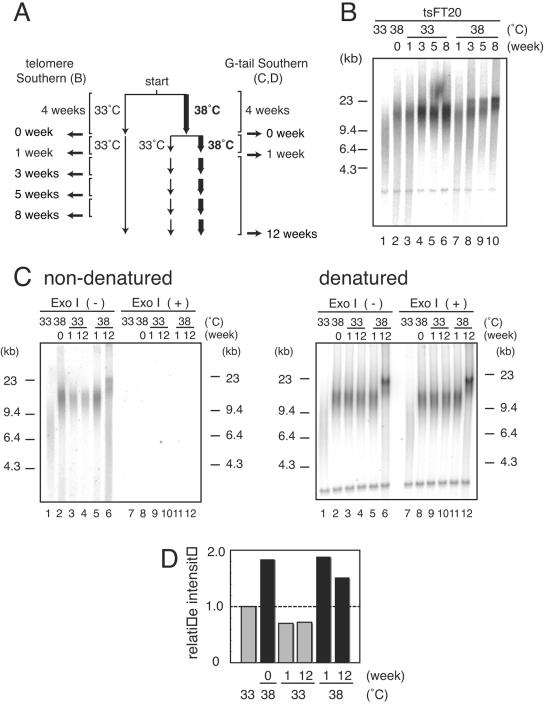
To determine whether the telomere elongation induced by impaired DNA polymerase α activity is reversible or not, we first prepared tsFT20 cells that had been cultured at 38°C for 4 weeks. The cultures were then either kept at 38°C or shifted to the permissive temperature of 33°C (week zero) and harvested 1, 3, 5, 8, or 12 weeks later, as shown in the protocol (Fig. 7A). tsFT20 cells at week zero showed telomere elongation, as expected (Fig. 7B, compare lane 1 with lane 2). When the cultures were continuously grown at 38°C for an additional 8 weeks, the telomere length was increased progressively during this period (lanes 7 to 10). In contrast, the cultures that were shifted to 33°C at week zero did not show further telomere lengthening (lanes 3 to 6). Interestingly, the telomeres in these cultures did not return to the original short size shown by those kept at 33°C. These results indicate that the elongation process per se is reversibly induced by the inactivation of DNA polymerase α activity. However, it appears that once the telomere length was altered by the 4-week culture at 38°C, the cells adapted to the new setting of telomere length and maintained the length even after DNA polymerase α activity was restored. In contrast, the G-tail extension that appeared at week zero was reduced to the original level as shown by tsFT20 cells cultured at 33°C for 1 week and for 12 weeks after shifting back to 33°C (Fig. 7C, lanes 2 to 4), suggesting that the G-tail length is directly related to DNA polymerase α activity.
FIG. 7.
Telomere length of tsFT20 cells that had been transiently cultured at semipermissive temperature. A. Protocol of the experiment. tsFT20 cells that had been cultured at 33°C or 38°C for 4 weeks were prepared (week zero). The 38°C cultures were divided into two: one was kept at 38°C, while the other was shifted to 33°C and cultured for an additional 12 weeks. The cells were harvested at the indicated time points and analyzed for telomere length as for Fig. 2. B. Telomere lengths of cells shown in panel A. C. G-tail signals of cells indicated in panel A. DNA samples were treated [Exo I (+)] or not treated [Exo I (-)] with Exo I prior to HinfI digestion. After in-gel hybridization and autoradiography (nondenatured), the gel was denatured and reprobed with the same telomeric oligonucleotide probe (denatured). D. The G-tail signal intensities obtained in panel C were quantitated and normalized to the signal intensity of the ∼4.0-kb internal telomeric band detected under denatured conditions. The value obtained for cells grown stably at 33°C is set at 1.0 (panel C, lane 1). Similar results were obtained from four independent experiments.
Chromosomal aberrations are accompanied by telomeric DNA and chromatin alterations in tsFT20 cells at semipermissive temperature.
Because we observed significantly altered structures of telomeric DNA and chromatin in tsFT20 cells at the semipermissive temperature, we investigated whether these alterations would have any impact on genome integrity. FM3A and tsFT20 cells were cultured at 33°C or 38°C for 6 weeks, and metaphase spreads were prepared from them. The spreads were analyzed by staining DNA as well as detecting telomeric repeats using FISH with the Cy3-conjugated (CCCTAA)3 PNA probe.
The Robertsonian (Rb) chromosome fusion refers to the centric fusion of two acrocentric chromosomes to form a single metacentric one (reviewed in reference 56). It is frequently observed in mouse genomes that are characterized by acrocentric chromosomes. We observed Rb fusions in all the 33°C or 38°C grown FM3A and tsFT20 cells we examined, indicating that FM3A cells had already acquired and stably transmitted some Rb fusions. In all cases, we did not detect the telomeric repeat signal at the fusion points in our cytological analyses. Interestingly, however, we found a significantly high frequency of cells containing large numbers of Rb fusions in tsFT20 cells cultured at 38°C for 6 weeks, compared with FM3A cells cultured at 33°C or 38°C and tsFT20 cells cultured at 33°C (Fig. 8A and B). When cells containing different numbers of Rb fusions were scored, tsFT20 cells cultured at 38°C displayed a marked increase in the percentage of Rb-containing cells: 88.2% of the cells contained more than five Rb fusions, whereas 2.8%, 3.1%, and 6.0% of the cells did so in FM3A at 33°C and 38°C and in tsFT20 cells cultured at 33°C, respectively (Fig. 8C). A significant fraction of tsFT20 cells cultured at 38°C contained six or more Rb fusions (52.9%), whereas none of the FM3A cells cultured at 33°C or 38°C, or tsFT20 cells cultured at 33°C, did so (Fig. 8D). Consistent with the increase in the number of Rb fusions, the total number of chromosomes in a cell was decreased in tsFT20 cells grown at 38°C (Fig. 8C). No telomeric repeat signals were observed at the fusion points in all the Rb fusion cases (Fig. 8A and B). This result was unexpected, given that the telomeric repeats were elongated rather than shortened in tsFT20 cells cultured at 38°C. We did not find any noticeably higher frequencies of chromatid fusions and breaks or of chromosome fusions and breaks. These results indicate that compromised DNA polymerase α activity entails a specific genomic instability in an unexpected manner, as revealed by the dramatic increases in the number of Rb fusions with no telomeric repeats at the fusion point.
DISCUSSION
In this study, we demonstrated that the compromised DNA polymerase α activity in tsFT20 cells leads to alterations in telomeric DNA and chromatin structures in a two-step manner. In the first step, which is evident 1 or 2 weeks after the temperature shift, G-tail extension occurs concomitantly with the accumulation of a large amount of POT1, but not of TRF1, at telomeres without overall telomeric repeat lengthening. Because this step is telomerase independent, it is most likely that the G-tail length is increased not by a net synthesis of the G-strand but by the exposure of the G-strand of preexisting telomeric double-stranded DNAs as the G-tail due to the erosion of the C-strand. The inefficient C-strand synthesis caused by the reduced DNA polymerase α primase activity during the conventional DNA replication may most likely explain the C-strand erosion. In the second step, the overall telomeric DNA length is progressively and markedly increased in a telomerase-dependent manner. At this stage, the amounts of both POT1 and TRF1 bound to telomeres are increased. These dynamic changes of telomeric DNAs and chromatin in polymerase α-deficient cells extend the observations in budding yeast and fission yeast polymerase α mutants (1, 2, 7, 12) and emphasize the evolutionarily conserved roles of DNA polymerase α in telomere maintenance. Unexpectedly, however, we observed a significantly high frequency of Rb fusions in tsFT20 cells at the semipermissive temperature. Thus, the hypomorphic function of DNA polymerase α results in rather specific chromosomal abnormalities.
Changes of abundance of POT1 and TRF1 at telomeres during G-tail length increase and telomeric DNA elongation.
We monitored the length of telomeric DNA along with the abundance of TRF1 and POT1 bound to telomeres during incubation of tsFT20 cells at 38°C. The telomeric proteins were analyzed by two independent experimental systems (quantitative ChIP and IF), which gave the same conclusions. Most significantly, a large absolute amount of POT1, but not of TRF1, associated with telomeres in the first step of telomeric DNA change, showing long single-stranded regions but not long double-stranded regions. In the second step, where both single-stranded and double-stranded telomeric DNA lengths were increased, both POT1 and TRF1 bound to telomeres in larger amounts than control cells. These results suggest that the absolute abundances of POT1 and TRF1 at telomeres are correlated with the lengths of single-stranded and double-stranded telomeric DNAs, respectively. Because POT1 is a DNA-binding protein specific to single-stranded telomeric DNA (5, 32, 37), this result appears to be explained by the larger amounts of POT1 recruited directly to longer single-stranded G-tails. However, recent studies have suggested that most POT1 proteins at telomeres are first recruited by protein-protein interactions with TPP1 (originally known as PIP1, PTOP, or TINT1) (35, 36, 68, 70) and thereafter loaded to the single-stranded regions. Therefore, the result presented in this study was not straightforwardly expected. It is believed that six telomeric proteins, TRF1, TRF2, Rap1, TIN2, TPP1, and POT1, comprise the core components of the mammalian telomeric complex (23, 34, 69). This study suggests that the stoichiometry of TRF1 and POT1 among these components is not strictly constant, but rather variable.
Mechanisms of telomeric elongation in tsFT20 cells at semipermissive temperature.
It has been reported that mutants of the catalytic subunit of DNA polymerase α show lengthening of telomeric DNAs in budding yeast and fission yeast (1, 2, 7, 12, 52). In most cases, the mutant alleles affect the catalytic activity of the enzyme, suggesting that the incomplete lagging strand synthesis at telomeres leads to structural changes of telomeric DNA and chromatin. In some cases, however, the mutant alleles deregulate telomere length in a more specific manner. Apparently, some budding yeast polymerase α mutants do not affect the catalytic activity of the enzyme; rather, they destabilize the protein-protein interaction between polymerase α and Cdc13 (52). These mutants show telomere elongation in a telomerase-dependent manner, suggesting that the protein interaction is involved in regulating telomerase action. A mutant allele of Pol12, the B subunit of budding yeast DNA polymerase α, was reported (19). The general DNA replication seems to be unaffected, whereas the specific interaction between Pol12 and Stn1, a telomere capping protein, is inactivated in the mutant. However, in these cases, telomere lengthening is relatively small and self-limited: the telomere size did not progressively increase; rather, it was stabilized in the new setting. In contrast to these specific alleles, telomere lengthening in catalytically defective mutants is characterized by massive and continuous elongation, as shown in tsFT20 cells. These differences in behavior among the polymerase α mutant alleles suggest that DNA polymerase α is involved in telomere maintenance via at least two distinct mechanisms: one is through the C-strand synthesis and the other is through protein-protein interactions with telomere-specific factors. We believe that the telomere phenotypes observed in tsFT20 cells were caused by the failure of general catalytic activity of DNA polymerase α.
Massive telomere elongation has been observed in ALT (alternative lengthening of telomeres) cells that maintain telomeres in a telomerase-independent manner using DNA recombination (reviewed in reference 47). However, we did not find any phenotypes that are closely associated with ALT, such as ALT-associated PML bodies (45, 71) (data not shown). Together with the finding that the elongation depends on telomerase activity, it is unlikely that the ALT mechanism is involved in the telomere elongation in tsFT20 cells.
Another example of massive telomere lengthening is the runaway telomere elongation found in the budding yeast Kluyveromyces lactis that harbors mutations in the template region of the telomerase RNA (42). We noticed that tsFT20 cells and K. lactis mutants share characteristic telomere phenotypes. First, these mutants showed more or less delays before the onset of telomere elongation. Second, the degree and the kinetics of telomere elongation were stochastic among cultures (Fig. 2; see also Fig. S1 in the supplemental material). Third, the elongation was progressive and massive (>10 kb in K. lactis mutants; telomere length in wild-type K. lactis is 0.25 to 0.5 kb) (Fig. 2) and continuous (at least 30 culture passages corresponding to approximately 1,000 generations in K. lactis mutants) (Fig. 2). It is believed that the substitution of mutant telomeric repeats for the wild-type repeats that took place during the initial lagging period disrupted the chromatin structure that inhibited telomerase action (42). Both double-stranded telomeric DNA-binding protein and G-tail-binding protein were implicated in the formation of the inhibitory chromatin. Once the “open” chromatin structure that is constitutively accessible to telomerase was formed, the runaway elongation of telomeres ensued.
We suspect that a similar mechanism operates in the continuous growth of telomeric DNA in tsFT20 cells at 38°C. It has been suggested that POT1 transduces the negative effect of TRF1 on telomerase action (36). In light of this, it is puzzling why telomeres kept elongated in tsFT20 cells at 38°C in the presence of large amounts of TRF1 and POT1 associated with telomeres. One possibility is that the TRF1-POT1 pathway does not simply inhibit telomerase action in proportion to the abundance of these proteins but positively and negatively regulates telomerase according to the state of the telomeric complex including TRF1 and POT1, which is reflected in the stoichiometry of the bound proteins in this study, as well as to the structures of telomeric DNA. The dynamic changes of the quantity and quality (such as protein modifications) of the telomeric proteins, as well as the modes of interactions among the protein components and telomeric DNAs, may intricately determine the accessibility of telomerase. In this sense, it is noteworthy that in our study there was a significant correlation between the increase in the length of the G-tail and the net increase of telomeric DNA by telomerase. Telomere elongation occurred only after the G-tail length was increased after shifting to the semipermissive temperature and stopped when the G-tail length returned to the control level after shifting from the semipermissive temperature to the permissive one. It is possible that the TRF1-POT1 pathway cannot regulate telomerase action when the length of G-tail is abnormally increased, as in cells under replication stress.
Although telomere homeostasis may be stabilized in a narrow range under stable conditions, the equilibrium is easily perturbed by genome stresses, such as inefficient DNA replication. We observed that tsFT20 cells that had been cultured at 38°C transiently for 4 weeks and then shifted to 33°C did not show any further increase or reduction of telomere length and appeared to have adapted to the new telomere length setting. tsFT20 cells that were shifted from 38°C to 33°C may maintain the altered telomeric chromatin even at 33°C and keep the equilibrium favorable for open chromatin. Alternatively, we cannot exclude the possibility that some genetic or epigenetic alteration(s) had occurred and been selected for growth advantage during the telomere elongation period of the cells, leading to the irreversible nature of the new state.
Chromosomal instability caused by polymerase α defects.
Because the p180 mutation allele present in tsFT20 cells affects the general catalytic activity of DNA polymerase α (Fig. 1), one may expect that genetic instability involving the whole genome would be observed in tsFT20 cells at the semipermissive temperature. This is indeed true, because it was previously found that tsFT20 cells that had been transiently cultured at the nonpermissive temperature (39°C) showed general and massive chromosomal aberrations, including chromatid gaps and breaks, chromosome pulverizations, and ring chromosomes (14). In contrast, we did not observe any significant increases of such chromosomal abnormalities other than Rb fusions in tsFT20 cells grown at the semipermissive temperature (38°C). These results indicate that the hypomorphic defects, rather than the complete inactivation, of DNA polymerase α manifest a rather specific spectrum of genetic instability.
Dicentric chromosomes were formed at a high incidence in human cells that had lost telomeric DNAs significantly after extensive propagation and entered the crisis stage (11). The disruption of telomere function by inactivating either telomerase or TRF proteins led to the frequent appearance of end-to-end fusions (6, 25, 30, 63). In mice, telomeres at short arms are generally shorter than those at long arms, making the short arm telomeres particularly vulnerable to these insults (72). Alternatively, Rb fusions have been observed frequently in mouse cells, simply because the other types of end-to-end fusions involving the telomeres of long chromosome arms result in the formation of very unstable dicentric chromosomes.
It is puzzling why chromosomes possessing long telomeric DNAs undergo Rb fusions with no telomeric FISH signal at the fusion points. Such cases are not unprecedented, however. Double-knockout mice defective in both the poly(ADP-ribose) polymerase gene (PARP) and the p53 gene showed very long telomeres as well as chromosomal abnormalities including Rb fusions. Significantly, the fusion point of Rb chromosome did not show telomeric FISH signals (61). In the PARP−/− p53−/− knockout mice, it was suggested that the telomeric DNA lengths were so heterogeneous, including very long and short ones, that some telomeres lacked telomeric DNA and were uncapped, leading to end-to-end fusions without telomeric DNA at the fusion points. Rapid telomere deletion, which was originally found in yeast and recently found in mammals (38), may have occurred in telomeres that underwent Rb fusion.
Alternatively, the Rb fusion observed in tsFT20 cells may not involve the failure of telomere capping. It has been reported that DNA polymerase α physically interacts with Swi6, an HP1 family heterochromatin protein that accumulates at telomeres, centromeres, and the silent mating type loci in fission yeast. A mutant allele of polymerase α dislocated Swi6 from these loci and derepressed gene silencing (3, 46). Because HP1 has been implicated in both centromeric and telomeric structures in metazoans (for examples, see references 18, 55, and 58), it is possible that the short arms of mouse cells, which contain mostly heterochromatin spanning from telomeres to centromeres, were disrupted to form the Rb fusions in polymerase α-defective cells. In this case, the Rb fusion was caused by the failure of maintenance of the mouse short arm heterochromatin and involved the fusion of such regions.
The specific increase in the incidence of Rb fusions in DNA polymerase α-compromised cells without general chromosomal instability, as well as the structural changes at telomere chromatin, supports the notion that telomeres and associated heterochromatin are loci that are particularly vulnerable to the failure of the DNA replication machinery. We have previously shown that the repetitive sequence of telomeric DNA per se is not a good substrate for conventional replication machinery in a simian virus 40 DNA-based in vitro replication system (48, 49). The binding of TRF1 and TRF2 to the sequence further increases the chance of replication stalling at telomeric repeats (48). Such conditions lead to significant alterations of the DNA and chromatin structures at telomeres, as shown in this study, which in turn may further deregulate telomere functions through, for example, unregulated telomerase actions. In this sense, telomeres should indeed be considered as the Achilles' heel of chromosomes, not only because of the end replication problem, as has been previously pointed out (20), but also as a locus where the tightly regulated replication machinery is essential for the maintenance of structure and function. It should be noted that the replication stress-induced deregulation of telomeres occurs in a relatively short period compared to that induced by the end replication problem. It is possible that similar conditions happen in cancer cells in vivo, thereby promoting the progression through genetic instability.
Supplementary Material
Acknowledgments
This work was supported by a COE Grant and Grants-in-Aid for Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology.
We are grateful to T. Kitamura (Institute of Medical Science, University of Tokyo) for pMX-puro and Platinum E cells. We thank Y. Shinkai (Institute for Virus Research, Kyoto University) for anti-mTRF1 antibody and critical reading of the manuscript, X. Yuan, R. Ohki, and R. Funayama for helpful discussions, M. Tamura for technical assistance, and A. Katayama, M. Sakamoto, M. Sasaki, and K. Fujimaki for excellent secretarial work.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, A. K., and C. Holm. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams Martin, A., I. Dionne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, S., S. Saini, S. Arora, and J. Singh. 2001. Chromodomain protein Swi6-mediated role of DNA polymerase alpha in establishment of silencing in fission yeast. J. Biol. Chem. 276:47814-47821. [DOI] [PubMed] [Google Scholar]
- 4.Armbruster, B. N., C. M. Linardic, T. Veldman, N. P. Bansal, D. L. Downie, and C. M. Counter. 2004. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 24:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 6.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 7.Carson, M. J., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 8.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 9.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colgin, L. M., K. Baran, P. Baumann, T. R. Cech, and R. R. Reddel. 2003. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 13:942-946. [DOI] [PubMed] [Google Scholar]
- 11.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahlen, M., P. Sunnerhagen, and T. S. Wang. 2003. Replication proteins influence the maintenance of telomere length and telomerase protein stability. Mol. Cell. Biol. 23:3031-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 14.Eki, T., T. Enomoto, Y. Murakami, F. Hanaoka, and M. Yamada. 1987. Characterization of chromosome aberrations induced by incubation at a restrictive temperature in the mouse temperature-sensitive mutant tsFT20 strain containing heat-labile DNA polymerase alpha. Cancer Res. 47:5162-5170. [PubMed] [Google Scholar]
- 15.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 16.Fan, X., and C. M. Price. 1997. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol. Biol. Cell 8:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, G., and T. R. Cech. 1995. Telomerase RNA localized in the replication band and spherical subnuclear organelles in hypotrichous ciliates. J. Cell Biol. 130:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanti, L., G. Giovinazzo, M. Berloco, and S. Pimpinelli. 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2:527-538. [DOI] [PubMed] [Google Scholar]
- 19.Grossi, S., A. Puglisi, P. V. Dmitriev, M. Lopes, and D. Shore. 2004. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 18:992-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 21.Herbert, B., A. E. Pitts, S. I. Baker, S. E. Hamilton, W. E. Wright, J. W. Shay, and D. R. Corey. 1999. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. USA 96:14276-14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hockemeyer, D., A. J. Sfeir, J. W. Shay, W. E. Wright, and T. de Lange. 2005. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 24:2667-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houghtaling, B. R., L. Cuttonaro, W. Chang, and S. Smith. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14:1621-1631. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, H. L., D. Gilley, E. H. Blackburn, and D. J. Chen. 1999. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 96:12454-12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwano, T., M. Tachibana, M. Reth, and Y. Shinkai. 2004. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 279:1442-1448. [DOI] [PubMed] [Google Scholar]
- 26.Izumi, M., H. Miyazawa, S. Harakawa, F. Yatagai, and F. Hanaoka. 1994. Identification of a point mutation in the cDNA of the catalytic subunit of DNA polymerase alpha from a temperature-sensitive mouse FM3A cell line. J. Biol. Chem. 269:7639-7644. [PubMed] [Google Scholar]
- 27.Kelleher, C., I. Kurth, and J. Lingner. 2005. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 25:808-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kipling, D., and H. J. Cooke. 1990. Hypervariable ultra-long telomeres in mice. Nature 347:400-402. [DOI] [PubMed] [Google Scholar]
- 29.Kitamura, T., M. Onishi, S. Kinoshita, A. Shibuya, A. Miyajima, and G. P. Nolan. 1995. Efficient screening of retroviral cDNA expression libraries. Proc. Natl. Acad. Sci. USA 92:9146-9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, H. W., M. A. Blasco, G. J. Gottlieb, J. W. Horner II, C. W. Greider, and R. A. DePinho. 1998. Essential role of mouse telomerase in highly proliferative organs. Nature 392:569-574. [DOI] [PubMed] [Google Scholar]
- 31.Lei, M., E. R. Podell, P. Baumann, and T. R. Cech. 2003. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426:198-203. [DOI] [PubMed] [Google Scholar]
- 32.Lei, M., E. R. Podell, and T. R. Cech. 2004. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11:1223-1229. [DOI] [PubMed] [Google Scholar]
- 33.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 34.Liu, D., M. S. O'Connor, J. Qin, and Z. Songyang. 2004. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279:51338-51342. [DOI] [PubMed] [Google Scholar]
- 35.Liu, D., A. Safari, M. S. O'Connor, D. W. Chan, A. Laegeler, J. Qin, and Z. Songyang. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6:673-680. [DOI] [PubMed] [Google Scholar]
- 36.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 37.Loayza, D., H. Parsons, J. Donigian, K. Hoke, and T. de Lange. 2004. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279:13241-13248. [DOI] [PubMed] [Google Scholar]
- 38.Lustig, A. J. 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4:916-923. [DOI] [PubMed] [Google Scholar]
- 39.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 40.Marcand, S., V. Brevet, C. Mann, and E. Gilson. 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10:487-490. [DOI] [PubMed] [Google Scholar]
- 41.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 42.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 43.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063-1066. [DOI] [PubMed] [Google Scholar]
- 44.Murakami, Y., H. Yasuda, H. Miyazawa, F. Hanaoka, and M. Yamada. 1985. Characterization of a temperature-sensitive mutant of mouse FM3A cells defective in DNA replication. Proc. Natl. Acad. Sci. USA 82:1761-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nabetani, A., O. Yokoyama, and F. Ishikawa. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 279:25849-25857. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama, J., R. C. Allshire, A. J. Klar, and S. I. Grewal. 2001. A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 20:2857-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neumann, A. A., and R. R. Reddel. 2002. Telomere maintenance and cancer—look, no telomerase. Nat. Rev. Cancer 2:879-884. [DOI] [PubMed] [Google Scholar]
- 48.Ohki, R., and F. Ishikawa. 2004. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 32:1627-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohki, R., T. Tsurimoto, and F. Ishikawa. 2001. In vitro reconstitution of the end replication problem. Mol. Cell. Biol. 21:5753-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poon, S. S., U. M. Martens, R. K. Ward, and P. M. Lansdorp. 1999. Telomere length measurements using digital fluorescence microscopy. Cytometry 36:267-278. [DOI] [PubMed] [Google Scholar]
- 51.Price, C. M. 1997. Synthesis of the telomeric C-strand. A review. Biochemistry (Moscow) 62:1216-1223. [PubMed] [Google Scholar]
- 52.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 53.Ray, S., Z. Karamysheva, L. Wang, D. E. Shippen, and C. M. Price. 2002. Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol. Cell. Biol. 22:5859-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 55.Sharma, G. G., K. K. Hwang, R. K. Pandita, A. Gupta, S. Dhar, J. Parenteau, M. Agarwal, H. J. Worman, R. J. Wellinger, and T. K. Pandita. 2003. Human heterochromatin protein 1 isoforms HP1(Hsα) and HP1(Hsβ) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol. Cell. Biol. 23:8363-8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slijepcevic, P. 1998. Telomeres and mechanisms of Robertsonian fusion. Chromosoma 107:136-140. [DOI] [PubMed] [Google Scholar]
- 57.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 58.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 59.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 60.Tatematsu, K., J. Nakayama, M. Danbara, S. Shionoya, H. Sato, M. Omine, and F. Ishikawa. 1996. A novel quantitative “stretch PCR assay” that detects a dramatic increase in telomerase activity during the progression of myeloid leukemias. Oncogene 13:2265-2274. [PubMed] [Google Scholar]
- 61.Tong, W. M., M. P. Hande, P. M. Lansdorp, and Z. Q. Wang. 2001. DNA strand break-sensing molecule poly(ADP-ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol. Cell. Biol. 21:4046-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 63.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 64.Veldman, T., K. T. Etheridge, and C. M. Counter. 2004. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 14:2264-2270. [DOI] [PubMed] [Google Scholar]
- 65.Wellinger, R. J., K. Ethier, P. Labrecque, and V. A. Zakian. 1996. Evidence for a new step in telomere maintenance. Cell 85:423-433. [DOI] [PubMed] [Google Scholar]
- 66.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 67.Wright, W. E., V. M. Tesmer, K. E. Huffman, S. D. Levene, and J. W. Shay. 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, Q., Y. L. Zheng, and C. C. Harris. 2005. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 25:1070-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye, J. Z., J. R. Donigian, M. van Overbeek, D. Loayza, Y. Luo, A. N. Krutchinsky, B. T. Chait, and T. de Lange. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279:47264-47271. [DOI] [PubMed] [Google Scholar]
- 70.Ye, J. Z., D. Hockemeyer, A. N. Krutchinsky, D. Loayza, S. M. Hooper, B. T. Chait, and T. de Lange. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]
- 72.Zijlmans, J. M., U. M. Martens, S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large interchromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94:7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.